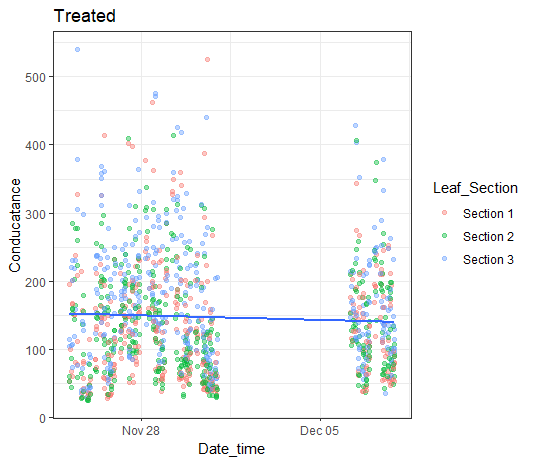
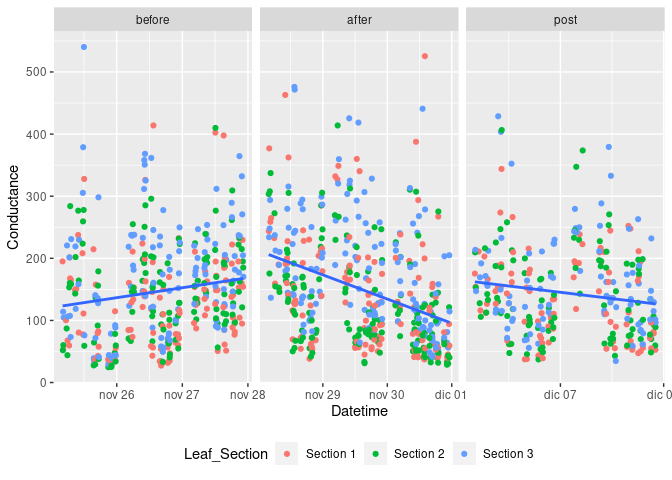
Here is a bit of a suggestion based on the data you supplied. Note I used {data.table} because it was convenient for me but you can do the same with base R or {tidyverse}.
library(patchwork)
library(data.table)
library(ggplot2)
## data in dput format below tho code.
mydat <- as.data.table(mydat)
mydat$Date_Time <- dmy_hm(mydat$Date_Time)
DD <- mydat[Date_Time > ymd("2022-11-30")]
DN <- mydat[Date_Time <= ymd("2022-11-30")]
pdec <- DD %>% ggplot(aes(Date_Time, Conductance)) + geom_point() +
geom_point(alpha=0.4, aes(colour= Leaf_Section)) +
ylim(0, 500) +
geom_smooth(method = lm, se = F) +
labs(x= "Date_time", y= "Conductance") +
theme(legend.position="none")
pnov <- DN %>% ggplot(aes(Date_Time, Conductance)) + geom_point()+
geom_point(alpha=0.4, aes(colour= Leaf_Section)) +
ylim(0, 500) +
geom_smooth(method = lm, se = F) +
labs(x= "Date_time", y= "Conductance")
pnov + pdec
######### data in data.frame layout#########################################################
mydat <- structure(list(Date_Time = c("25-Nov-22 4:10 AM", "25-Nov-22 4:23 AM", "25-Nov-22 8:44 AM", "25-Nov-22 9:11 AM", "25-Nov-22 3:28 PM", "25-Nov-22 3:40 PM",
"25-Nov-22 8:50 PM", "25-Nov-22 9:01 PM", "26-Nov-22 4:16 AM", "26-Nov-22 4:27 AM", "26-Nov-22 9:22 AM", "26-Nov-22 9:28 AM", "26-Nov-22 12:57 PM",
"26-Nov-22 1:01 PM", "26-Nov-22 4:11 PM", "26-Nov-22 4:15 PM", "26-Nov-22 6:35 PM", "26-Nov-22 6:41 PM", "26-Nov-22 9:31 PM", "26-Nov-22 9:36 PM",
"27-Nov-22 5:29 AM", "27-Nov-22 5:37 AM", "27-Nov-22 8:59 AM", "27-Nov-22 9:05 AM", "27-Nov-22 12:52 PM", "27-Nov-22 12:56 PM", "27-Nov-22 3:56 PM",
"27-Nov-22 4:01 PM", "27-Nov-22 7:06 PM", "27-Nov-22 7:11 PM", "27-Nov-22 10:02 PM", "27-Nov-22 10:14 PM", "28-Nov-22 7:30 AM", "28-Nov-22 7:45 AM",
"28-Nov-22 10:00 AM", "28-Nov-22 10:07 AM", "28-Nov-22 12:40 PM", "28-Nov-22 12:45 PM", "28-Nov-22 3:40 PM", "28-Nov-22 3:47 PM", "28-Nov-22 6:53 PM",
"28-Nov-22 7:01 PM", "28-Nov-22 9:18 PM", "28-Nov-22 9:31 PM", "29-Nov-22 4:38 AM", "29-Nov-22 8:17 AM", "29-Nov-22 8:24 AM", "29-Nov-22 12:01 PM",
"29-Nov-22 12:05 PM", "29-Nov-22 3:26 PM", "29-Nov-22 3:31 PM", "29-Nov-22 6:06 PM", "29-Nov-22 6:10 PM", "29-Nov-22 8:29 PM", "29-Nov-22 8:34 PM",
"30-Nov-22 3:48 AM", "30-Nov-22 3:55 AM", "30-Nov-22 9:31 AM", "30-Nov-22 9:35 AM", "30-Nov-22 11:04 AM", "30-Nov-22 11:08 AM", "30-Nov-22 1:32 PM",
"30-Nov-22 1:34 PM", "30-Nov-22 4:08 PM", "30-Nov-22 4:12 PM", "30-Nov-22 6:27 PM", "30-Nov-22 6:30 PM", "30-Nov-22 10:12 PM", "30-Nov-22 10:15 PM",
"06-Dec-22 4:59 AM", "06-Dec-22 5:25 AM", "06-Dec-22 9:20 AM", "06-Dec-22 9:27 AM", "06-Dec-22 11:59 AM", "06-Dec-22 12:07 PM", "06-Dec-22 4:06 PM",
"06-Dec-22 4:12 PM", "06-Dec-22 6:47 PM", "06-Dec-22 6:52 PM", "06-Dec-22 9:45 PM", "06-Dec-22 9:50 PM", "07-Dec-22 4:17 AM", "07-Dec-22 4:28 AM",
"07-Dec-22 10:07 AM", "07-Dec-22 10:13 AM", "07-Dec-22 12:08 PM", "07-Dec-22 12:14 PM", "07-Dec-22 4:16 PM", "07-Dec-22 4:19 PM", "07-Dec-22 6:35 PM",
"07-Dec-22 6:39 PM", "07-Dec-22 9:37 PM", "07-Dec-22 9:41 PM", "25-Nov-22 4:16 AM", "25-Nov-22 4:26 AM", "25-Nov-22 8:47 AM", "25-Nov-22 8:55 AM",
"25-Nov-22 3:34 PM", "25-Nov-22 3:41 PM", "25-Nov-22 8:56 PM", "25-Nov-22 9:05 PM", "26-Nov-22 4:19 AM", "26-Nov-22 4:30 AM"),
Plant_ID = c("C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2",
"C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2",
"C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2",
"C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2",
"C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2"),
Leaf.A.B = c("a", "b", "a", "b", "a", "b", "a", "b", "b", "a", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "b", "a", "a", "b", "a", "b", "a",
"b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b",
"a", "b", "a", "b", "b", "a", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b",
"a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "a", "b", "b", "a"),
Leaf_Section = c("Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1",
"Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1",
"Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1",
"Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1",
"Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1",
"Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1",
"Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1",
"Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1",
"Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1", "Section 1",
"Section 1", "Section 1", "Section 1", "Section 2", "Section 2", "Section 2", "Section 2", "Section 2", "Section 2", "Section 2",
"Section 2", "Section 2", "Section 2"),
Conductance = c(195.1, 53.3, 149.2, 150.8, 214.6, 29.9, 37.1, 31.3, 84.5, 160.4, 223.8, 121.7, 145.8, 43.1, 54.1, 27.3, 56.7, 31.8, 64.3, 78, 134.4,
163.6, 118.8, 162.9, 148.8, 50.8, 164.4, 51.3, 77, 81.5, 229.7, 154.1, 190, 151.2, 463, 191, 116.3, 69.6, 71.5, 54.3, 230.1, 38.3, 68.1,
81.3, 332, 168.9, 128.9, 55.7, 57.9, 87.8, 45, 53.7, 56.8, 50.3, 74.2, 129.9, 98.2, 78.4, 51, 48.1, 45.8, 39.6, 42.4, 40.7, 40.3, 143.3,
39.8, 34, 41.2, 211, 116.1, 188.1, 152.9, 127.4, 61.4, 152.8, 37.7, 98.5, 43.4, 106.5, 103.4, 192.3, 238.6, 210.2, 112.3, 136.2, 66, 247.8,
66.8, 194.1, 50.2, 81.3, 79.1, 52, 60.2, 143.2, 202, 27.6, 64.5, 24.7, 26.8, 70.7, 70.9)), class = "data.frame", row.names = c(NA, -103L))