New to r and shiny, wrote my first app to show colleagues some healthcare related data -
Whilst it works perfectly laptop, it seems to only generate some of the plots when deployed
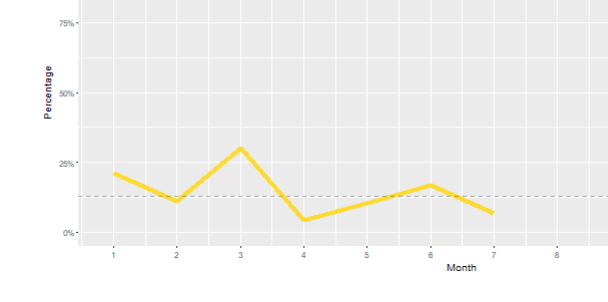
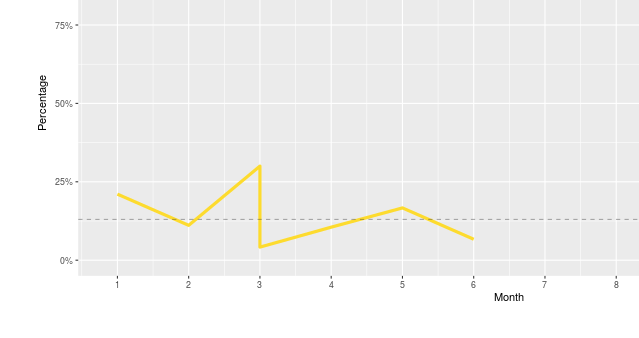
There was no error message, some of the plots were blank but another using the same data looks fine.
library(tidyverse)
library(shiny)
library(lubridate)
ERAS <- readRDS("ERAS.rda")
ERAS_yearly <- ERAS %>%
arrange(year_m) %>%
mutate(DoSdate = ymd(year_m), yearDoS = year(DoSdate), monthDoS = month(DoSdate)) %>%
group_by(yearDoS) %>%
summarise(year_meanLOS = mean(Postop_LOS),
year_meanAll3 = mean(All_3 == T, na.rm = T),
year_AKI = mean(AKI_pos == T, na.rm = T))
# User Interface
ui <- fluidPage(
titlePanel("NERCI Data"),
sidebarLayout(
sidebarPanel(
print("Choose the following for graphs on Tabs 1 and 2:"),
sliderInput("year", "Select Year:", animate = T,
min = 2016, max = 2019, value = 2016, sep = ""),
checkboxGroupInput("approach", "Surgical Approach", c("Laparoscopic", "Lap-assisted", "Converted to open", "Open"),
selected = c("Laparoscopic", "Lap-assisted", "Converted to open", "Open")),
print("Choose the following for graph on Tab 3:"),
selectInput("x_axis", "Choose x axis",
c("Surgical Risk" = "Risk",
"Obesity Class" = "Obesity",
"ASA-PS" = "ASA",
"Anaemia" = "Anaemia",
"Blood Transfusion" = "BTS",
"Acute Kidney Injury" = "AKI_pos",
"DrEaming Goals" = "All_3",
"Surgical Approach" = "Sx_Approach",
"POMS on Day 3" = "posPOMS3",
"POMS on Day 5" = "posPOMS5",
"Re-operation for Leaks" = "Reoperation_Leak",
"Re-operation for Other" = "Reoperation_Other",
"ICU Admission" = "ICU_Admission"),
selected = "Surgical Risk"),
selectInput("y_axis", "Choose y axis",
c("Total Post-op Length of Stay" = "Postop_LOS",
"SHDU Length of Stay" = "HDU_LOS")),
selectInput("extra", "Choose additional info",
c("Surgical Approach" = "Sx_Approach",
"Surgical Risk" = "Risk",
"Obesity Class" = "Obesity"),
selected = "Surgical Approach"),
dateRangeInput("date", "Choose Date Range of Data to display",
start = "2016-02-01", end = Sys.Date(),
min = "2016-02-01", max = Sys.Date())
),
mainPanel(
tabsetPanel(
tabPanel("DrEaMing",
plotOutput(outputId = "LOS_plot"),
plotOutput(outputId = "All3_plot")),
tabPanel("AKI Data",
plotOutput(outputId = "AKI_plot")),
tabPanel("Length of Stay Data",
plotOutput(outputId = "LOS_overall"))
)
)
)
)
# Server
server <- function(input, output){
output$LOS_plot <- renderPlot({
plot1a <- ERAS %>% filter(Sx_Approach %in% input$approach) %>%
arrange(year_m) %>%
mutate(DoSdate = ymd(year_m), yearDoS = year(DoSdate), monthDoS = month(DoSdate)) %>%
filter(yearDoS == input$year) %>%
group_by(year_m) %>%
mutate(mean_LOS = mean(Postop_LOS)) %>%
ggplot(aes(x = monthDoS, y = mean_LOS)) + geom_line(colour = "#A8659C", size = 1.5) +
geom_hline(data = ERAS_yearly[ERAS_yearly$yearDoS == input$year, ], aes(yintercept = year_meanLOS), alpha = 0.4, colour = "#323232", linetype = "dashed") +
scale_x_continuous(limits = c(1, 12), breaks = c(1:12)) +
scale_y_continuous(limits = c(0,30), minor_breaks = 1) +
theme(aspect.ratio = 0.4) + xlab("Month") + ylab("Days") + ggtitle("Mean Length of Stay over Time")
plot1a
})
output$All3_plot <- renderPlot({
plot1b <- ERAS %>% filter(Sx_Approach %in% input$approach) %>%
arrange(year_m) %>%
mutate(DoSdate = ymd(year_m), yearDoS = year(DoSdate), monthDoS = month(DoSdate)) %>%
filter(yearDoS == input$year) %>% group_by(year_m) %>%
mutate(mean_All3 = mean(All_3 == T, na.rm = T)) %>%
ggplot(aes(x = monthDoS, y = mean_All3)) + geom_line(colour = "#116133", size = 1.5) +
geom_hline(data = ERAS_yearly[ERAS_yearly$yearDoS == input$year, ], aes(yintercept = year_meanAll3), alpha = 0.4, colour = "#323232", linetype = "dashed") +
scale_x_continuous(limits = c(1, 12), breaks = c(1:12)) +
scale_y_continuous(labels = scales::percent, limits = c(0, 1)) +
theme(aspect.ratio = 0.4) + xlab("Month") + ylab("Percentage") + ggtitle("Patients Achieving All Three ERAS Goals")
plot1b
})
output$AKI_plot <- renderPlot({
plot2 <- ERAS %>% filter(Sx_Approach %in% input$approach) %>%
arrange(year_m) %>%
mutate(DoSdate = ymd(year_m), yearDoS = year(DoSdate), monthDoS = month(DoSdate)) %>%
filter(yearDoS == input$year) %>% group_by(year_m) %>%
mutate(mean_AKI = mean(AKI_pos == T, na.rm = T)) %>%
ggplot(aes(x = monthDoS, y = mean_AKI)) + geom_line(colour = "#FDDB2E", size = 1.5) +
geom_hline(data = ERAS_yearly[ERAS_yearly$yearDoS == input$year, ], aes(yintercept = year_AKI), alpha = 0.4, colour = "#323232", linetype = "dashed") +
scale_x_continuous(limits = c(1, 12), breaks = c(1:12)) +
scale_y_continuous(labels = scales::percent, limits = c(0, 1)) +
theme(aspect.ratio = 0.4) + xlab("Month") + ylab("Percentage") + ggtitle("Percentage of Patients with Post-op AKI")
plot2
})
output$LOS_overall <- renderPlot({
x <- input$x_axis
y <- input$y_axis
extra <- input$extra
ERAS <- filter(ERAS, DoS >= input$date[1] & DoS <= input$date[2])
plot3 <- ggplot(data = ERAS, aes_string(x = x, y = y)) +
geom_jitter(aes_string(colour = extra), alpha = 0.5, size = 5) +
geom_boxplot(alpha = 0.5) +
ggtitle("Length of Stay Post-op") + xlab(x) + ylab("Length of Stay") +
theme(aspect.ratio = 0.6)
plot3
})
}
shinyApp(ui = ui, server = server)
I've looked for solutions online which seem to suggest checking whether the versions are the same but being new to coding, I have no idea how to approach this.
(Edit: I have included the code for the app now.)
System Information:
- RStudio Edition: (Desktop)
- RStudio Version: 1.2.5001
- OS Version: Windows 10
- R Version: 3.6.1
-
sessionInfo():
Referred here from support.rstudio.com