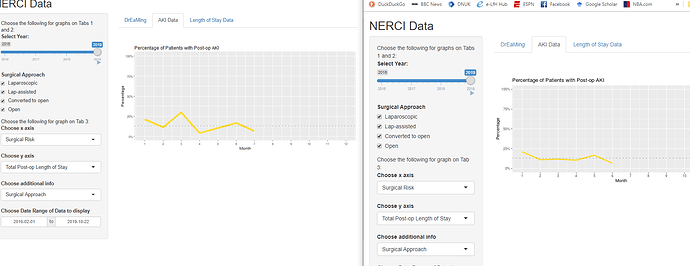
I deployed my first app on shinyapps.io but noticed that the plots are behaving differently when compared to running the app on my machine (the more accurate plots are produced locally).
This is the code I have for the app:
library(tidyverse)
library(shiny)
ERAS <- readRDS("ERAS.rda")
ERAS$year_m2 <- as.character(ERAS$year_m)
ERAS_yearly <- ERAS %>%
arrange(year_m) %>%
mutate(monthDoS = as.numeric(substr(year_m2, 6, 7)), yearDoS = as.numeric(substr(year_m2, 1, 4))) %>%
group_by(yearDoS) %>%
summarise(year_meanLOS = mean(Postop_LOS),
year_meanAll3 = mean(All_3 == T, na.rm = T),
year_AKI = mean(AKI_pos == T, na.rm = T))
# User Interface
ui <- fluidPage(
titlePanel("NERCI Data"),
sidebarLayout(
sidebarPanel(
print("Choose the following for graphs on Tabs 1 and 2:"),
sliderInput("year", "Select Year:", animate = T,
min = 2016, max = 2019, value = 2016, sep = ""),
checkboxGroupInput("approach", "Surgical Approach", c("Laparoscopic", "Lap-assisted", "Converted to open", "Open"),
selected = c("Laparoscopic", "Lap-assisted", "Converted to open", "Open")),
print("Choose the following for graph on Tab 3:"),
selectInput("x_axis", "Choose x axis",
c("Surgical Risk" = "Risk",
"Obesity Class" = "Obesity",
"ASA-PS" = "ASA",
"Anaemia" = "Anaemia",
"Blood Transfusion" = "BTS",
"Acute Kidney Injury" = "AKI_pos",
"DrEaming Goals" = "All_3",
"Surgical Approach" = "Sx_Approach",
"POMS on Day 3" = "posPOMS3",
"POMS on Day 5" = "posPOMS5",
"Re-operation for Leaks" = "Reoperation_Leak",
"Re-operation for Other" = "Reoperation_Other",
"ICU Admission" = "ICU_Admission"),
selected = "Surgical Risk"),
selectInput("y_axis", "Choose y axis",
c("Total Post-op Length of Stay" = "Postop_LOS",
"SHDU Length of Stay" = "HDU_LOS")),
selectInput("extra", "Choose additional info",
c("Surgical Approach" = "Sx_Approach",
"Surgical Risk" = "Risk",
"Obesity Class" = "Obesity"),
selected = "Surgical Approach"),
dateRangeInput("date", "Choose Date Range of Data to display",
start = "2016-02-01", end = Sys.Date(),
min = "2016-02-01", max = Sys.Date())
),
mainPanel(
tabsetPanel(
tabPanel("DrEaMing",
plotOutput(outputId = "LOS_plot"),
plotOutput(outputId = "All3_plot")),
tabPanel("AKI Data",
plotOutput(outputId = "AKI_plot")),
tabPanel("Length of Stay Data",
plotOutput(outputId = "LOS_overall"))
)
)
)
)
# Server
server <- function(input, output){
output$LOS_plot <- renderPlot({
ERAS <- ERAS %>%
arrange(year_m) %>%
mutate(monthDoS = as.numeric(substr(year_m2, 6, 7)), yearDoS = as.numeric(substr(year_m2, 1, 4))) %>%
filter(yearDoS == input$year) %>% filter(Sx_Approach %in% input$approach) %>%
group_by(monthDoS) %>%
mutate(mean_LOS = mean(Postop_LOS))
plot1a <- ggplot(data = ERAS, aes(x = monthDoS, y = mean_LOS)) + geom_line(colour = "#A8659C", size = 1.5) +
geom_hline(data = ERAS_yearly[ERAS_yearly$yearDoS == input$year, ], aes(yintercept = year_meanLOS), alpha = 0.4, colour = "#323232", linetype = "dashed") +
scale_x_continuous(limits = c(1, 12), breaks = c(1:12)) +
scale_y_continuous(limits = c(0,30), minor_breaks = 1) +
theme(aspect.ratio = 0.4) + xlab("Month") + ylab("Days") + ggtitle("Mean Length of Stay over Time")
plot1a
})
output$All3_plot <- renderPlot({
ERAS <- ERAS %>%
arrange(year_m) %>%
mutate(monthDoS = as.numeric(substr(year_m2, 6, 7)), yearDoS = as.numeric(substr(year_m2, 1, 4))) %>%
filter(yearDoS == input$year) %>% filter(Sx_Approach %in% input$approach) %>%
group_by(monthDoS) %>%
mutate(mean_All3 = mean(All_3 == T, na.rm = T))
plot1b <- ggplot(data = ERAS, aes(x = monthDoS, y = mean_All3)) + geom_line(colour = "#116133", size = 1.5) +
geom_hline(data = ERAS_yearly[ERAS_yearly$yearDoS == input$year, ], aes(yintercept = year_meanAll3), alpha = 0.4, colour = "#323232", linetype = "dashed") +
scale_x_continuous(limits = c(1, 12), breaks = c(1:12)) +
scale_y_continuous(labels = scales::percent, limits = c(0, 1)) +
theme(aspect.ratio = 0.4) + xlab("Month") + ylab("Percentage") + ggtitle("Patients Achieving All Three ERAS Goals")
plot1b
})
output$AKI_plot <- renderPlot({
ERAS <- ERAS %>%
arrange(year_m) %>%
mutate(monthDoS = as.numeric(substr(year_m2, 6, 7)), yearDoS = as.numeric(substr(year_m2, 1, 4))) %>%
filter(yearDoS == input$year) %>% filter(Sx_Approach %in% input$approach) %>%
group_by(monthDoS) %>%
mutate(mean_AKI = mean(AKI_pos == T, na.rm = T))
plot2 <- ggplot(data = ERAS, aes(x = monthDoS, y = mean_AKI)) + geom_line(colour = "#FDDB2E", size = 1.5) +
geom_hline(data = ERAS_yearly[ERAS_yearly$yearDoS == input$year, ], aes(yintercept = year_AKI), alpha = 0.4, colour = "#323232", linetype = "dashed") +
scale_x_continuous(limits = c(1, 12), breaks = c(1:12)) +
scale_y_continuous(labels = scales::percent, limits = c(0, 1)) +
theme(aspect.ratio = 0.4) + xlab("Month") + ylab("Percentage") + ggtitle("Percentage of Patients with Post-op AKI")
plot2
})
output$LOS_overall <- renderPlot({
x <- input$x_axis
y <- input$y_axis
extra <- input$extra
ERAS <- filter(ERAS, DoS >= input$date[1] & DoS <= input$date[2])
plot3 <- ggplot(data = ERAS, aes_string(x = x, y = y)) +
geom_jitter(aes_string(colour = extra), alpha = 0.5, size = 5) +
geom_boxplot(alpha = 0.5) +
ggtitle("Length of Stay Post-op") + xlab(x) + ylab("Length of Stay") +
theme(aspect.ratio = 0.6)
plot3
})
}
shinyApp(ui = ui, server = server)
The plot that is accurate is the one on the left in the attached file. It seems that something is happening consistently in the month of March and April on the server version. It is also somehow missing a month.
This is the
dput()
dput(test2019)
structure(list(Sx_Approach = structure(c(1L, 1L, 2L, 1L, 1L,
1L, 1L, 3L, 4L, 1L, 1L, 1L, 4L, 4L, 2L, 1L, 1L, 1L, 1L, 4L, 4L,
4L, 1L, 1L, 2L, 1L, 1L, 4L, 3L, 4L, 1L, 1L, 1L, 4L, 1L, 2L, 4L,
1L, 1L, 1L, 1L, 4L, 3L, 1L, 1L, 4L, 1L, 1L, 2L, 4L, 2L, 4L, 1L,
4L, 4L, 1L, 1L, 4L, 1L, 4L, 1L, 4L, 4L, 4L, 4L, 1L, 2L, 1L, 1L,
4L, 4L, 1L, 1L, 1L, 3L, 4L, 4L, 4L, 4L, 1L, 1L, 1L, 4L, 1L, 1L,
1L, 1L, 4L, 1L, 1L, 1L, 1L, 1L, 1L, 3L, 4L, 1L, 4L, 1L, 3L, 4L,
1L, 3L, 1L, 4L, 1L, 3L, 1L, 2L, 4L, 1L, 1L, 3L, 1L, 4L, 4L, 4L,
4L, 2L, 1L, 1L, 1L, 1L), .Label = c("Laparoscopic", "Lap-assisted",
"Converted to open", "Open"), class = "factor"), mean_LOS = c(8.80392156862745,
8.80392156862745, 8.80392156862745, 8.80392156862745, 8.80392156862745,
8.80392156862745, 8.80392156862745, 8.80392156862745, 8.80392156862745,
8.80392156862745, 8.80392156862745, 8.80392156862745, 8.80392156862745,
8.80392156862745, 8.80392156862745, 8.80392156862745, 8.80392156862745,
8.80392156862745, 8.80392156862745, 7.20833333333333, 7.20833333333333,
7.20833333333333, 7.20833333333333, 7.20833333333333, 7.20833333333333,
7.20833333333333, 7.20833333333333, 7.20833333333333, 7.20833333333333,
7.20833333333333, 7.20833333333333, 7.20833333333333, 7.20833333333333,
7.20833333333333, 7.20833333333333, 7.20833333333333, 7.20833333333333,
7.02, 7.02, 7.02, 7.02, 7.02, 7.02, 7.02, 7.02, 7.02, 7.02, 8.12962962962963,
8.12962962962963, 8.12962962962963, 8.12962962962963, 8.12962962962963,
8.12962962962963, 8.12962962962963, 8.12962962962963, 8.12962962962963,
8.12962962962963, 8.12962962962963, 8.12962962962963, 8.12962962962963,
8.12962962962963, 8.12962962962963, 8.12962962962963, 8.12962962962963,
8.12962962962963, 8.12962962962963, 8.12962962962963, 8.12962962962963,
8.12962962962963, 8.12962962962963, 8.12962962962963, 8.03703703703704,
8.03703703703704, 8.03703703703704, 8.03703703703704, 8.03703703703704,
8.03703703703704, 8.03703703703704, 8.03703703703704, 8.03703703703704,
8.03703703703704, 8.03703703703704, 8.03703703703704, 8.03703703703704,
8.03703703703704, 8.03703703703704, 8.03703703703704, 8.03703703703704,
8.03703703703704, 8.03703703703704, 8.43076923076923, 8.43076923076923,
8.43076923076923, 8.43076923076923, 8.43076923076923, 8.43076923076923,
8.43076923076923, 8.43076923076923, 8.43076923076923, 8.43076923076923,
8.43076923076923, 8.43076923076923, 8.43076923076923, 8.43076923076923,
8.43076923076923, 8.43076923076923, 8.43076923076923, 8.43076923076923,
6.01639344262295, 6.01639344262295, 6.01639344262295, 6.01639344262295,
6.01639344262295, 6.01639344262295, 6.01639344262295, 6.01639344262295,
6.01639344262295, 6.01639344262295, 6.01639344262295, 6.01639344262295,
6.01639344262295, 6.01639344262295, 6.01639344262295), monthDoS = c(1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3,
3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4,
4, 4, 4, 4, 4, 4, 4, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5,
5, 5, 5, 5, 5, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6,
6, 6, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7), yearDoS = c(2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019), mean_aki = c(0.210526315789474, 0.210526315789474, 0.210526315789474,
0.210526315789474, 0.210526315789474, 0.210526315789474, 0.210526315789474,
0.210526315789474, 0.210526315789474, 0.210526315789474, 0.210526315789474,
0.210526315789474, 0.210526315789474, 0.210526315789474, 0.210526315789474,
0.210526315789474, 0.210526315789474, 0.210526315789474, 0.210526315789474,
0.111111111111111, 0.111111111111111, 0.111111111111111, 0.111111111111111,
0.111111111111111, 0.111111111111111, 0.111111111111111, 0.111111111111111,
0.111111111111111, 0.111111111111111, 0.111111111111111, 0.111111111111111,
0.111111111111111, 0.111111111111111, 0.111111111111111, 0.111111111111111,
0.111111111111111, 0.111111111111111, 0.3, 0.3, 0.3, 0.3, 0.3,
0.3, 0.3, 0.3, 0.3, 0.3, 0.0416666666666667, 0.0416666666666667,
0.0416666666666667, 0.0416666666666667, 0.0416666666666667, 0.0416666666666667,
0.0416666666666667, 0.0416666666666667, 0.0416666666666667, 0.0416666666666667,
0.0416666666666667, 0.0416666666666667, 0.0416666666666667, 0.0416666666666667,
0.0416666666666667, 0.0416666666666667, 0.0416666666666667, 0.0416666666666667,
0.0416666666666667, 0.0416666666666667, 0.0416666666666667, 0.0416666666666667,
0.0416666666666667, 0.0416666666666667, 0.105263157894737, 0.105263157894737,
0.105263157894737, 0.105263157894737, 0.105263157894737, 0.105263157894737,
0.105263157894737, 0.105263157894737, 0.105263157894737, 0.105263157894737,
0.105263157894737, 0.105263157894737, 0.105263157894737, 0.105263157894737,
0.105263157894737, 0.105263157894737, 0.105263157894737, 0.105263157894737,
0.105263157894737, 0.166666666666667, 0.166666666666667, 0.166666666666667,
0.166666666666667, 0.166666666666667, 0.166666666666667, 0.166666666666667,
0.166666666666667, 0.166666666666667, 0.166666666666667, 0.166666666666667,
0.166666666666667, 0.166666666666667, 0.166666666666667, 0.166666666666667,
0.166666666666667, 0.166666666666667, 0.166666666666667, 0.0666666666666667,
0.0666666666666667, 0.0666666666666667, 0.0666666666666667, 0.0666666666666667,
0.0666666666666667, 0.0666666666666667, 0.0666666666666667, 0.0666666666666667,
0.0666666666666667, 0.0666666666666667, 0.0666666666666667, 0.0666666666666667,
0.0666666666666667, 0.0666666666666667)), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"), row.names = c(NA, -123L), groups = structure(list(
monthDoS = c(1, 2, 3, 4, 5, 6, 7), .rows = list(1:19, 20:37,
38:47, 48:71, 72:90, 91:108, 109:123)), row.names = c(NA,
-7L), class = c("tbl_df", "tbl", "data.frame")))
Any comments as to why this would occur would be greatly appreciated.
Follow-up: I checked the logs on the app and noticed the following warnings:
Failed to create bus connection: No such file or directory
Warning in system("timedatectl", intern = TRUE) :
running command 'timedatectl' had status 1
Are they related to my problem?