Hello all ![]()
I have been trying to self-study different statistical concepts and have now found a use case that could be a relevant learning opportunity - calculating sample sizes for an experimental design.
Background
I have a marketing activity that I would like to test. The activity consists of an email campaign sent to a number of recipients with an intent to buy. What I'm recording is whether or not a recipient makes a purchase (purchase: yes / no).
In order to understand the effect of sending this email (that is, the proportion of recipients contacted that makes a purchase out of all recipients contacted - a conversion rate), I would like to hold out a group of customers who should not be contacted - a control group.
The question is: how many of my total group size should I (randomly) allocate to the control group?
I have some information from previous trials (note, these segments are independent - no one recipient is in both segment A and B):
| Year | Segment | Treatment (contacted) group size | Control (not contacted) group size | Conversion % for treatment | Conversion % for control |
|------|---------|----------------------------------|------------------------------------|----------------------------|--------------------------|
| 2022 | A | 40000 | 2000 | 0.014 (1.4 %) | 0.009 (0.9 %) |
| 2022 | B | 7000 | 200 | 0.007 (0.7 %) | 0.0029 (0.29 %) |
As I understand the design, in order to estimate the adequacy of this historical trial, I need to work with a two-sample test for proportions for unequal sample sizes.
I found the pwr package and tried testing the above historical studies by running the following code (assuming a significance level of 0.05 and a required power of 0.8):
# Seed
set.seed(42)
# libraries
library(pwr)
# Segment A
## Effect size
(effect_size_segment_A <- pwr::ES.h(p1 = 0.014, p2 = 0.009))
#> [1] 0.04717644
## Power calculation for two proportions (different sample sizes)
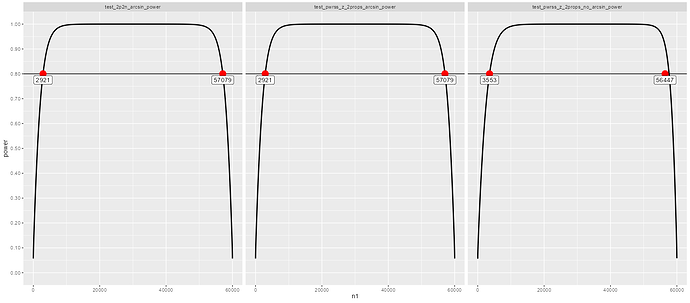
(pwr::pwr.2p2n.test(h = effect_size_segment_A, n1 = 40000, n2 = 2000, power = 0.8, sig.level = NULL, alternative = "two.sided"))
#>
#> difference of proportion power calculation for binomial distribution (arcsine transformation)
#>
#> h = 0.04717644
#> n1 = 40000
#> n2 = 2000
#> sig.level = 0.2227702
#> power = 0.8
#> alternative = two.sided
#>
#> NOTE: different sample sizes
# Segment B
## Effect size
(effect_size_segment_B <- pwr::ES.h(p1 = 0.007, p2 = 0.0029))
#> [1] 0.05977242
## Power calculation for two proportions (different sample sizes)
(pwr::pwr.2p2n.test(h = effect_size_segment_B, n1 = 7000, n2 = 200, power = 0.8, sig.level = NULL, alternative = "two.sided"))
#>
#> difference of proportion power calculation for binomial distribution (arcsine transformation)
#>
#> h = 0.05977242
#> n1 = 7000
#> n2 = 200
#> sig.level = 0.7209852
#> power = 0.8
#> alternative = two.sided
#>
#> NOTE: different sample sizes
Created on 2023-11-08 with reprex v2.0.2
If my understanding and code is correct, it seems like the previous design for both trials were underpowered.
New trials
Now that I want to send out a new campaign activity, I have the chance of controlling the number of recipients that I should allocate to the control (or holdout) group. Below I have total segment size and effect size assumptions (in this case the assumed effect sizes are equal to those experienced in the previous trial):
| Year | Segment | Total segment size | Treatment (contacted) group size | Control (not contacted) group size | Assumed conversion % for treatment | Assumed conversion % for control |
|------|---------|--------------------|----------------------------------|------------------------------------|------------------------------------|----------------------------------|
| 2023 | A | 60000 | ? | ? | 0.014 (1.4 %) | 0.009 (0.9 %) |
| 2023 | B | 4500 | ? | ? | 0.007 (0.7 %) | 0.0029 (0.29 %) |
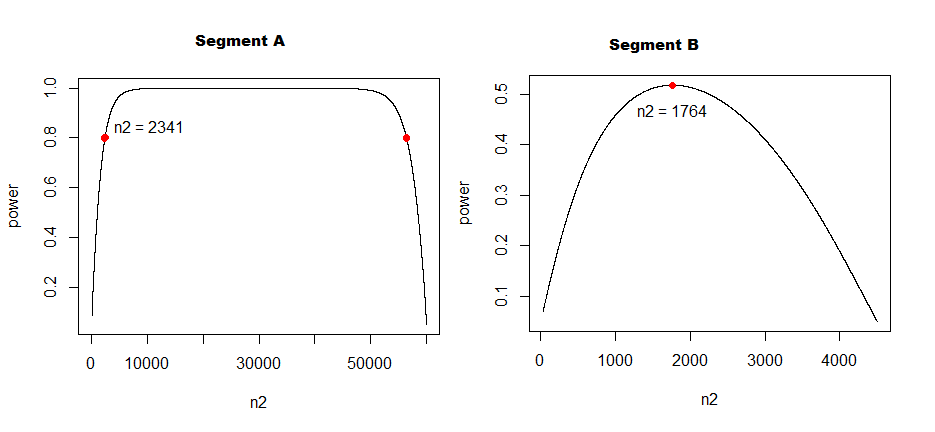
Following the logic from above, and assuming the same realised effect size as in the historical trial (the two proportions within each segment not falling any closer to each), I have ended up calculating the control group size like in the following:
# Seed
set.seed(42)
# libraries
library(pwr)
# Segment A
## Effect size
(effect_size_segment_A <- pwr::ES.h(p1 = 0.014, p2 = 0.009))
#> [1] 0.04717644
## Power calculation for two proportions (different sample sizes)
(pwr::pwr.2p2n.test(h = effect_size_segment_A, n1 = 56000, power = 0.8, sig.level = 0.05, alternative = "two.sided"))
#>
#> difference of proportion power calculation for binomial distribution (arcsine transformation)
#>
#> h = 0.04717644
#> n1 = 56000
#> n2 = 3763.614
#> sig.level = 0.05
#> power = 0.8
#> alternative = two.sided
#>
#> NOTE: different sample sizes
# Segment B
## Effect size
(effect_size_segment_B <- pwr::ES.h(p1 = 0.007, p2 = 0.0029))
#> [1] 0.05977242
(pwr::pwr.2p2n.test(h = effect_size_segment_B, n1 = 4500, power = 0.8, sig.level = 0.05, alternative = "two.sided"))
#>
#> difference of proportion power calculation for binomial distribution (arcsine transformation)
#>
#> h = 0.05977242
#> n1 = 4500
#> n2 = 4292.393
#> sig.level = 0.05
#> power = 0.8
#> alternative = two.sided
#>
#> NOTE: different sample sizes
Created on 2023-11-08 with reprex v2.0.2
The above results seem reasonable, although it is obvious that segment B total size is hopelessly small.
Now to my main question:
- Is this the correct statistical approach? Should I instead work with a chi sq test and a 2 x 2 matrix of the counts - if so, I would very much appreciate some guidance.
Regards