I am working on a Rmd script. This script includes a code chunk for sql:
{r packages, echo=TRUE}
library(tidyverse)
library(rmarkdown)
library(dbplyr)
library(DBI)
library(odbc)
con <- dbConnect(odbc(), "Athena")
some_var <- 7
some_other_var <- 30
Then I get my data via a sql code chunk:
{sql eval=FALSE, connection=con, include=FALSE, output.var="rawd"}
select *
from table
where blah <= ?some_var
The sql chunk uses variables declared further up in the script, in this case some_var referenced in the sql with ?some_var.
That returns a new variable 'rawd' which allows me to move on to reprocessing...
{r preprocessing, echo=TRUE, message=FALSE, warning=FALSE, cache=F}
processed_data <- rawd %>% ... do stuff
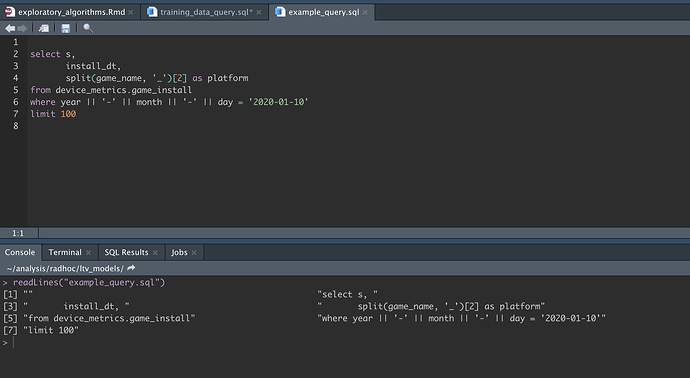
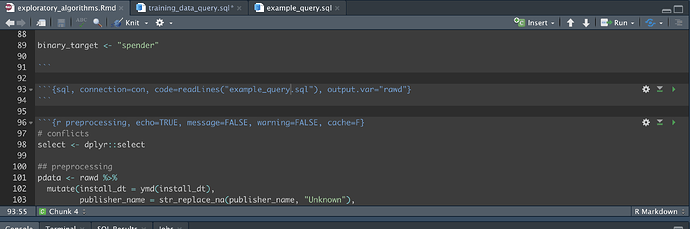
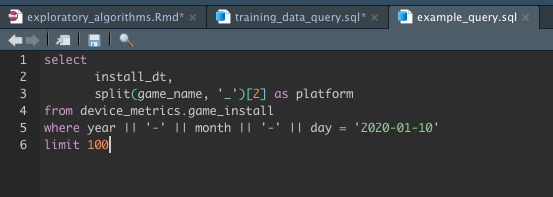
But actually, my SQL chunk is large and although I can minimize it with the small expander arrows to the left of the code text, I'd like to move it into it's own file. So I clicked on the new file icon and selected sql script then pasted the sql in there.
But, I have two problems:
-
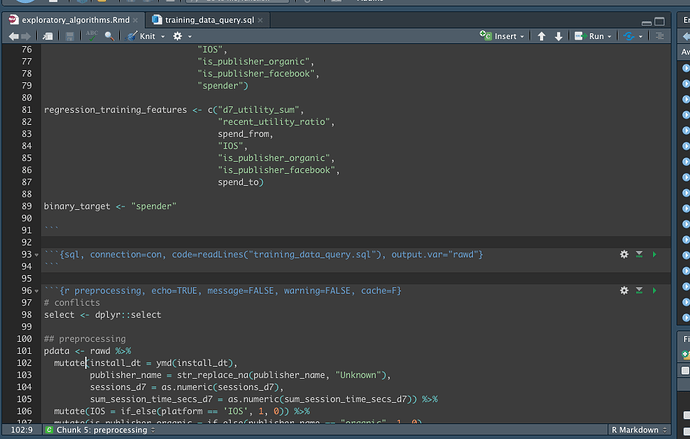
On the current sql chunk I can have the output of the query stored in a variable with
output.var="rawd", resulting in a new df called 'rawd'. If I import the new sql file, called 'training_data_query.sql', how can I tell the parent file, the working Rmd file to store the results as 'rawd'? I.e. What's the prescribed way to source a .sql file from within a Rmd file? -
In the example above I reference variables within the sql chunk by prepending the variable name with a question mark e.g
where blah <= ?some_var. How can I do that when the sql is it's own file rather than a sql chunk?
Desired script structure:
{r packages, echo=TRUE}
library(tidyverse)
library(rmarkdown)
library(dbplyr)
library(DBI)
library(odbc)
con <- dbConnect(odbc(), "Athena")
some_var <- 7
some_other_var <- 30
{r preprocessing, echo=TRUE, message=FALSE, warning=FALSE, cache=F}
source(training_data.sql) # ideally results in a new df called 'rawd'
processed_data <- rawd %>% ... do stuff