Consider the following code block:
library(dplyr)
library(dbplyr)
iris_db <- tbl_memdb(iris)
con <- src_memdb()$con
query <- "select * from iris where Species = 'setosa'"
setosa <- dbGetQuery(con, query)
If I run this, I get a data frame back with just the setosa rows from iris. So this works fine.
However, I'd like to include some variables within my script e.g.
species <- 'setosa'
query2 <- paste0("select * from iris where Species = '", species, "'")
setosa2 <- dbGetQuery(con, query2)
This also works, a new df setosa2 is returned just like with my first attempt where I hard coded the species to be returned.
However, in my actual script there are many variables I would like to use within the sql query, so it would get unsightly and hard to read the sql query after not too long.
I really enjoy working with .sql files within rstudio and recently posted a similar question within the context of integrating a .sql file and .Rmd file.
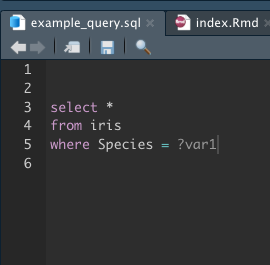
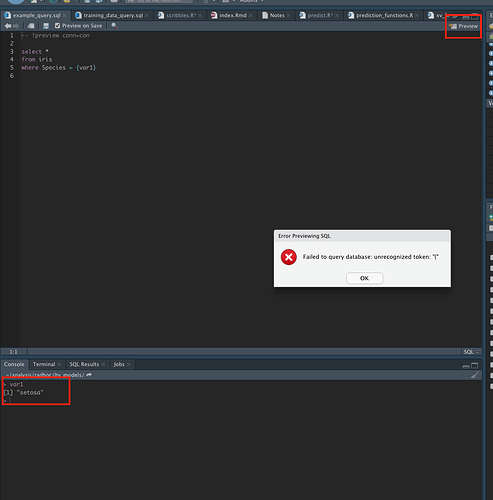
Within an Rmd file one can refer to a sql code chunk and integrate variables using question marks e.g.
```{sql connection=con, include=FALSE, output.var="my_df"}
select * from iris where Species = ?species
```
My question is, is there a workflow where I can integrate an external sql script within to a r script (as opposed to a Rmd) file? With the Rmd file one can use readLines but I think that only works when calling knit.
Or must I just use paste() with a long string and variables to generate my query and pass to r?