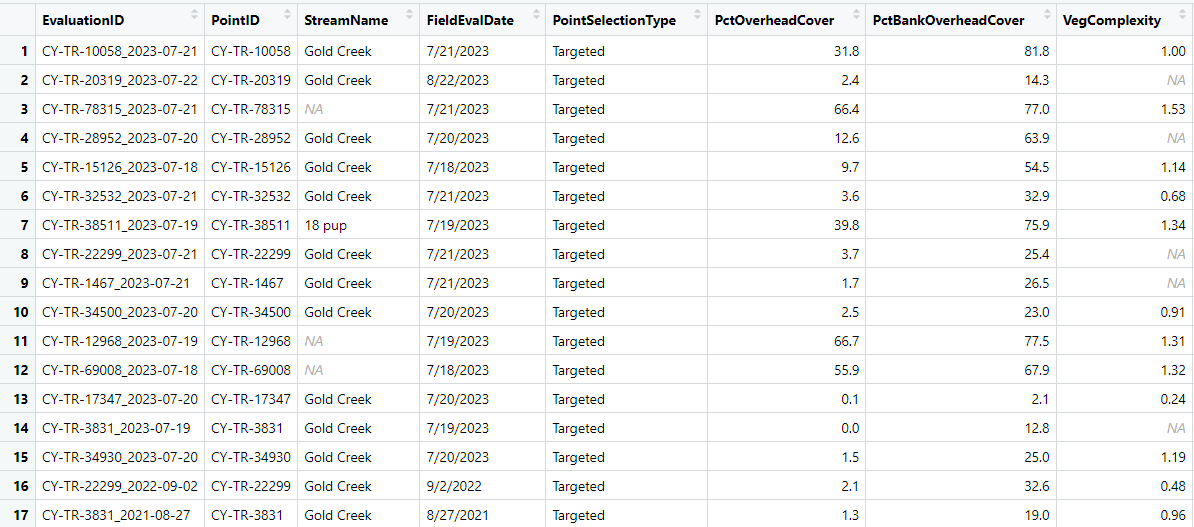
Here is a example of the data.frame where I no longer have Two separate dataframes but all data is in one dataframe.
data.frame(
stringsAsFactors = FALSE,
EvaluationID = c("CY-TR-10058_2023-07-21",
"CY-TR-20319_2023-07-22","CY-TR-78315_2023-07-21",
"CY-TR-28952_2023-07-20","CY-TR-15126_2023-07-18",
"CY-TR-32532_2023-07-21","CY-TR-38511_2023-07-19",
"CY-TR-22299_2023-07-21",
"CY-TR-1467_2023-07-21","CY-TR-34500_2023-07-20",
"CY-TR-12968_2023-07-19","CY-TR-69008_2023-07-18",
"CY-TR-17347_2023-07-20",
"CY-TR-3831_2023-07-19","CY-TR-34930_2023-07-20",
"CY-TR-22299_2022-09-02","CY-TR-3831_2021-08-27",
"CY-TR-28952_2021-08-28","CY-TR-20319_2021-08-28",
"CY-TR-1467_2021-08-29"),
PointID = c("CY-TR-10058","CY-TR-20319","CY-TR-78315",
"CY-TR-28952","CY-TR-15126","CY-TR-32532",
"CY-TR-38511","CY-TR-22299","CY-TR-1467",
"CY-TR-34500","CY-TR-12968","CY-TR-69008",
"CY-TR-17347","CY-TR-3831","CY-TR-34930",
"CY-TR-22299","CY-TR-3831","CY-TR-28952",
"CY-TR-20319","CY-TR-1467"),
StreamName = c("Gold Creek","Gold Creek",NA,"Gold Creek",
"Gold Creek","Gold Creek","18 pup",
"Gold Creek","Gold Creek","Gold Creek",NA,NA,
"Gold Creek","Gold Creek","Gold Creek",
"Gold Creek","Gold Creek","Gold Creek",
"Gold Creek","Gold Creek"),
FieldEvalDate = c("7/21/2023","8/22/2023","7/21/2023",
"7/20/2023","7/18/2023","7/21/2023","7/19/2023",
"7/21/2023","7/21/2023","7/20/2023",
"7/19/2023","7/18/2023","7/20/2023","7/19/2023",
"7/20/2023","9/2/2022","8/27/2021",
"8/28/2021","8/28/2021","8/29/2021"),
PointSelectionType = c("Targeted","Targeted","Targeted","Targeted",
"Targeted","Targeted","Targeted",
"Targeted","Targeted","Targeted","Targeted",
"Targeted","Targeted","Targeted","Targeted",
"Targeted","Targeted","Targeted",
"Targeted","Targeted"),
PctOverheadCover = c(31.8,
2.4,66.4,12.6,9.7,3.6,39.8,3.7,1.7,
2.5,66.7,55.9,0.1,0,1.5,2.1,1.3,10.6,
0,2.3),
PctBankOverheadCover = c(81.8,
14.3,77,63.9,54.5,32.9,75.9,25.4,
26.5,23,77.5,67.9,2.1,12.8,25,32.6,19,
41.4,4.1,25.4),
VegComplexity = c(1,NA,
1.53,NA,1.14,0.68,1.34,NA,NA,0.91,
1.31,1.32,0.24,NA,1.19,0.48,0.96,0.75,
0.45,0.68),
VegComplexityWoody = c(0.76,
NA,0.71,NA,0.96,0.45,0.81,NA,NA,
0.68,0.67,0.66,0.14,NA,0.69,0.36,0.44,
0.32,0.27,0.3),
VegComplexityUnderstoryGround = c(0.59,
NA,0.43,NA,0.86,0.43,0.79,NA,NA,
0.58,0.67,0.63,0.14,NA,0.68,0.34,0.43,
0.3,0.27,0.29),
SpecificConductance = c(723,
NA,1522,NA,663,861,299.1,NA,NA,829,
402,547,670,NA,755,664.6,670.1,805.2,
774.9,832.4),
pH = c(8.29,
NA,8.32,NA,7.72,8.16,7.6,NA,NA,7.99,
8.3,8.25,8.22,NA,7.94,8.24,8.05,
8.11,8.28,8.37),
InstantTemp = c(9,NA,
8.7,NA,6.33,7.46,5.8,NA,NA,11.8,
2.5,6.6,7.8,NA,5.86,6.9,2.4,0.4,3.5,
3.9),
TurbidityAvg = c(6.54,
NA,3.46,NA,37.2,18.53,22.6,NA,NA,
19.82,22.9,579,43.47,NA,9.65,28.64,5.75,
15.06,25.94,13.77),
PoolCount = c(3,NA,
15,NA,2,11,12,NA,NA,8,33,9,1,NA,
2,1,1,2,1,0),
PctPools = c(5.57,
NA,11.6,NA,5.87,11.62,26.67,NA,NA,
18.24,26.25,9.37,1.73,NA,5.21,3.42,3,
2.67,2.68,0)
)
#> EvaluationID PointID StreamName FieldEvalDate
#> 1 CY-TR-10058_2023-07-21 CY-TR-10058 Gold Creek 7/21/2023
#> 2 CY-TR-20319_2023-07-22 CY-TR-20319 Gold Creek 8/22/2023
#> 3 CY-TR-78315_2023-07-21 CY-TR-78315 <NA> 7/21/2023
#> 4 CY-TR-28952_2023-07-20 CY-TR-28952 Gold Creek 7/20/2023
#> 5 CY-TR-15126_2023-07-18 CY-TR-15126 Gold Creek 7/18/2023
#> 6 CY-TR-32532_2023-07-21 CY-TR-32532 Gold Creek 7/21/2023
#> 7 CY-TR-38511_2023-07-19 CY-TR-38511 18 pup 7/19/2023
#> 8 CY-TR-22299_2023-07-21 CY-TR-22299 Gold Creek 7/21/2023
#> 9 CY-TR-1467_2023-07-21 CY-TR-1467 Gold Creek 7/21/2023
#> 10 CY-TR-34500_2023-07-20 CY-TR-34500 Gold Creek 7/20/2023
#> 11 CY-TR-12968_2023-07-19 CY-TR-12968 <NA> 7/19/2023
#> 12 CY-TR-69008_2023-07-18 CY-TR-69008 <NA> 7/18/2023
#> 13 CY-TR-17347_2023-07-20 CY-TR-17347 Gold Creek 7/20/2023
#> 14 CY-TR-3831_2023-07-19 CY-TR-3831 Gold Creek 7/19/2023
#> 15 CY-TR-34930_2023-07-20 CY-TR-34930 Gold Creek 7/20/2023
#> 16 CY-TR-22299_2022-09-02 CY-TR-22299 Gold Creek 9/2/2022
#> 17 CY-TR-3831_2021-08-27 CY-TR-3831 Gold Creek 8/27/2021
#> 18 CY-TR-28952_2021-08-28 CY-TR-28952 Gold Creek 8/28/2021
#> 19 CY-TR-20319_2021-08-28 CY-TR-20319 Gold Creek 8/28/2021
#> 20 CY-TR-1467_2021-08-29 CY-TR-1467 Gold Creek 8/29/2021
#> PointSelectionType PctOverheadCover PctBankOverheadCover VegComplexity
#> 1 Targeted 31.8 81.8 1.00
#> 2 Targeted 2.4 14.3 NA
#> 3 Targeted 66.4 77.0 1.53
#> 4 Targeted 12.6 63.9 NA
#> 5 Targeted 9.7 54.5 1.14
#> 6 Targeted 3.6 32.9 0.68
#> 7 Targeted 39.8 75.9 1.34
#> 8 Targeted 3.7 25.4 NA
#> 9 Targeted 1.7 26.5 NA
#> 10 Targeted 2.5 23.0 0.91
#> 11 Targeted 66.7 77.5 1.31
#> 12 Targeted 55.9 67.9 1.32
#> 13 Targeted 0.1 2.1 0.24
#> 14 Targeted 0.0 12.8 NA
#> 15 Targeted 1.5 25.0 1.19
#> 16 Targeted 2.1 32.6 0.48
#> 17 Targeted 1.3 19.0 0.96
#> 18 Targeted 10.6 41.4 0.75
#> 19 Targeted 0.0 4.1 0.45
#> 20 Targeted 2.3 25.4 0.68
#> VegComplexityWoody VegComplexityUnderstoryGround SpecificConductance pH
#> 1 0.76 0.59 723.0 8.29
#> 2 NA NA NA NA
#> 3 0.71 0.43 1522.0 8.32
#> 4 NA NA NA NA
#> 5 0.96 0.86 663.0 7.72
#> 6 0.45 0.43 861.0 8.16
#> 7 0.81 0.79 299.1 7.60
#> 8 NA NA NA NA
#> 9 NA NA NA NA
#> 10 0.68 0.58 829.0 7.99
#> 11 0.67 0.67 402.0 8.30
#> 12 0.66 0.63 547.0 8.25
#> 13 0.14 0.14 670.0 8.22
#> 14 NA NA NA NA
#> 15 0.69 0.68 755.0 7.94
#> 16 0.36 0.34 664.6 8.24
#> 17 0.44 0.43 670.1 8.05
#> 18 0.32 0.30 805.2 8.11
#> 19 0.27 0.27 774.9 8.28
#> 20 0.30 0.29 832.4 8.37
#> InstantTemp TurbidityAvg PoolCount PctPools
#> 1 9.00 6.54 3 5.57
#> 2 NA NA NA NA
#> 3 8.70 3.46 15 11.60
#> 4 NA NA NA NA
#> 5 6.33 37.20 2 5.87
#> 6 7.46 18.53 11 11.62
#> 7 5.80 22.60 12 26.67
#> 8 NA NA NA NA
#> 9 NA NA NA NA
#> 10 11.80 19.82 8 18.24
#> 11 2.50 22.90 33 26.25
#> 12 6.60 579.00 9 9.37
#> 13 7.80 43.47 1 1.73
#> 14 NA NA NA NA
#> 15 5.86 9.65 2 5.21
#> 16 6.90 28.64 1 3.42
#> 17 2.40 5.75 1 3.00
#> 18 0.40 15.06 2 2.67
#> 19 3.50 25.94 1 2.68
#> 20 3.90 13.77 0 0.00
Created on 2024-02-06 with reprex v2.1.0
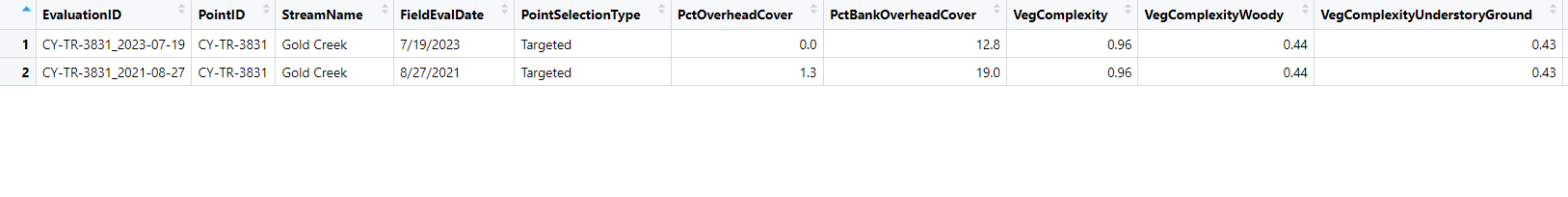
As an example of what I would like to happen: Look at rows 14 and 17 ($PointID for both those rows is CY-TR-3831). You will notice the values for the rows in $PointID are the same (this is the unique identifier). Then look at the $FieldEvalDate, you will see row 17 is 8/27/2021 and row 14 is 7/19/2023. I would like to over-write row 17 column values with that of row 14 (thus "updating" the row 17 with data from 14). As an example this should result in row 17 $PctBankOverheadCover changing from 19 to 12.8, but for $VegComplexity the value in row 17 is unchanged because the value in row 14 is NA.
I do not want to update ALL the columns therefore I tried to create a vector where the elements are column names and I thought I could feed this into my filter which is what I tried to do in the original post with two data.frames.
I hope this provides some clarity on what I am trying to accomplish. This is the thread where I got the idea to make two data.frames
Merge Dataframes to Fill Missing Data in RStudio - #5 by andresrcs