Good day!
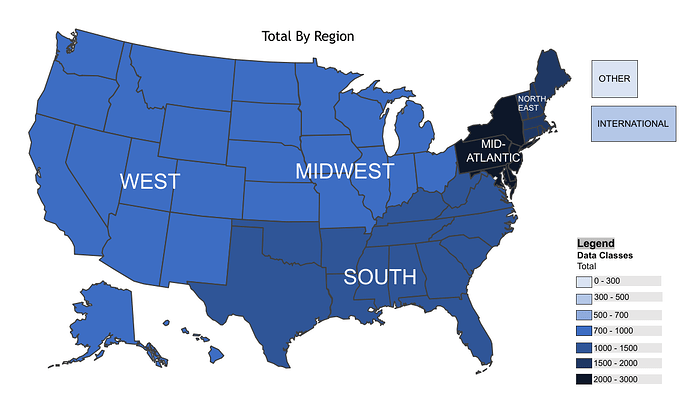
I would like to create a choropleth map with R using USA regions as the boundaries. Here is an example created in PowerPoint:
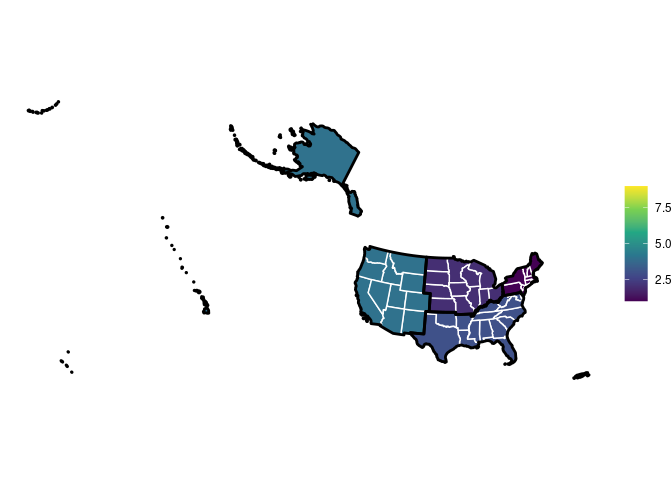
This is what I could acchieve with qmplot:
library(tidyverse)
library(ggmap)
library(ggrepel)
data <- tibble::tribble(
~lon, ~lat,
-98.133208, 11.4326077,
-89.552784, 11.634501,
-62.2766186, 44.5090949,
-65.3279894, 33.1067754,
-67.7095365, 44.6294348,
-96.552784, 35.634501,
-97.3279093, 29.7724417,
-82.6363869, 28.2949194,
-80.2061931, 46.0133808,
-72.014118, 32.4681642,
-76.2531465, 47.3666368,
-82.1650991, 46.7758541,
-5.696645, 11.945587,
-112.707349, 38.5205043,
-63.0884036, 52.3930959,
-87.128901, 39.1242719,
-65.1626756, 31.3463503,
-94.3254958, 40.3274999,
-98.56121, 42.5770056,
-115.4429944, 46.1502862,
-117.7901088, 30.6913751,
-63.7389596, 54.6584068,
-109.1147095, 24.2156978,
-119.8340735, 22.6832497,
-117.8780275, 37.7311394,
-67.1763467, 38.5861576,
-96.4427769, 25.14644,
-78.0814292, 15.5386936,
-74.4185584, 34.5834425,
-77.4185584, 36.5834425,
-79.4185584, 37.5834425,
-63.121085, 33.9241038,
-88.121085, 41.9241038,
-112.7260713, 40.836309,
-90.552784, 30.634501,
-109.552784, 12.634501,
-73.9224329, 48.6153549
)
data <-
data %>% sample_n(200, replace = TRUE) %>% mutate(
total = round( runif(nrow(.), min = 100, max = 10000), digits = 0) ,
lon = lon + round( runif(nrow(.), min = -10, max = 10), digits = 0),
lat = lat + round( runif(nrow(.), min = -10, max = 10), digits = 0)
)
qmplot(x = lon, y = lat, data = data, maptype = "toner-lite", legend = "topleft" , alpha = .3,geom = c("point","density2d"), size = total)+ theme(legend.position = "none")+
geom_label_repel(aes(label=total),
box.padding = 0.35,
point.padding = 0.5,
segment.color = 'grey50')
#> Using zoom = 3...
#> Map tiles by Stamen Design, under CC BY 3.0. Data by OpenStreetMap, under ODbL.
#> Warning in min(x): no non-missing arguments to min; returning Inf
#> Warning in max(x): no non-missing arguments to max; returning -Inf
#> Warning in min(x): no non-missing arguments to min; returning Inf
#> Warning in max(x): no non-missing arguments to max; returning -Inf
#> Warning: ggrepel: 174 unlabeled data points (too many overlaps). Consider
#> increasing max.overlaps
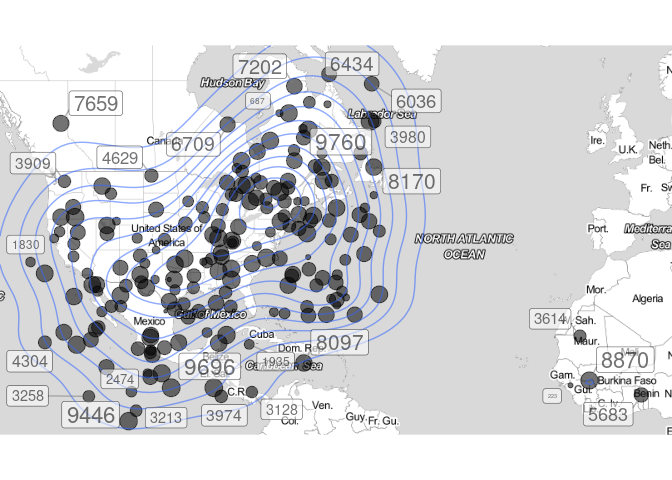
Created on 2021-09-16 by the reprex package (v2.0.1)
Any suggestions?
Thank you very much.
André