I am using RMarkdown to create a pdf report and I need the graphs and tables to appear in the order in which they are generated and placed within the text at the point where they are generated in the Rmardown file. This seeems like a reasonable thing to want to do. However...strange - the plots all go to the end and out of order. Why? How can I make them show up in the order of execution in the Rmarkdown document??
NOTE: when I generate the report as a HTML or a word doc, the tables and figures appear in the correct order. But I need to prepare the report as a pdf and would like to do that directly.
I have seen other similar questions asked here, but the level of discussion is over my head and it is not clear to me that anyone has resolved this problem. Some of the discussion involves Tex commands that I am not familiar with. I know there is an issue with "float" and I have read what is available on the web, but I am new to Rmarkdown/Bookdown and really cannot interpret the information. Perhaps it is not possible to do what I want to do and if so I would appreciate knowing that. but if it is possible I would be eternally grateful if someone can show me how to modify this Rmarkdown file to make that happen.... Thanks!!!
FYI I am running this file under RStudio Version 1.2.1335, which is running R version 3.6.0.
The "offending" Rmd file is given below that should execute and exhibit the strange behavior described above...
---
title: "My title"
author: "Me"
output:
word_document: default
extra_dependencies: float
fig_caption: yes
html_document:
df_print: paged
number_sections: yes
pdf_document: default
bookdown::pdf_document2: null
---
```{r}
knitr::opts_chunk$set(fig.pos = "!H")
```
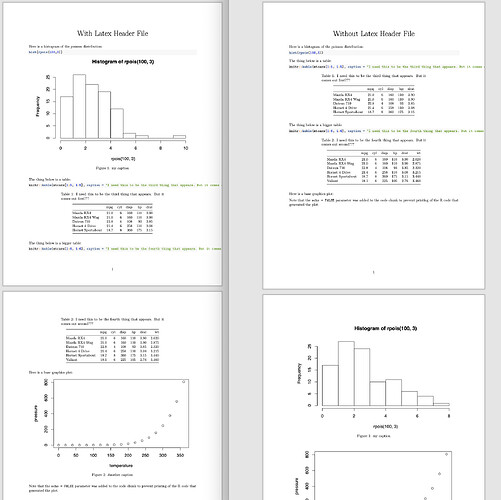
```{r plot1,fig.cap="This graph is supposed to appear first, but it comes out third???",fig.height = 5, fig.width = 5}
x <- 1:10
y <- rnorm(10)
plot(x,y)
```
```{r plot2, fig.cap="This graph is supposed to appear second but it comes out last???.",fig.height = 5, fig.width = 5}
plot(y,x)
```
The thing below is a table
```{r table1}
knitr::kable(mtcars[1:5, 1:5], caption = "I need this to be the third thing that appears. But it comes out first???")
```
The thing below is a bigger table
```{r table2}
knitr::kable(mtcars[1:6, 1:6], caption = "I need this to be the fourth thing that appears. But it comes out second???")
```