Packages needing compilation, such as brms, need writing rights for the /tmp directory or they will issue an error (see https://forum.posit.co/t/will-rstudio-cloud-permit-packages-that-need-compilation/20883).
On my local machine, I solve the problem by running sudo mount -o remount, exec /tmp in the terminal. However, to do that you need super user rights, which is not the case in RStudio Cloud. Any other solution?
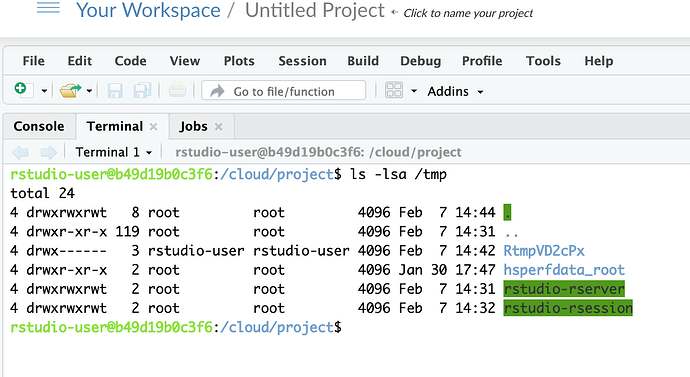
/tmp is writeable, there should be no need to remount it.
Could you share a specific case where you are experiencing an issue?
This is the output of a small script I use for teaching when running on RSrudio Cloud. The same problem is solved on my Xubuntu 18.04 by allowing to write and execute files in the /tmp directory (sorry I forgot to mention that before, although it was clear from the shell command) . Thank you!
library(brms)
Loading required package: Rcpp
Loading required package: ggplot2
Loading 'brms' package (version 2.7.0). Useful instructions
can be found by typing help('brms'). A more detailed introduction
to the package is available through vignette('brms_overview').
Run theme_set(theme_default()) to use the default bayesplot theme.
dat <- read.csv("SocInfExperiment.csv")
dat$status <- relevel(dat$status, ref="low")
summary(dat)
distance social.influence status
Min. :1.970 no :148 low :191
1st Qu.:2.690 yes:153 high:110
Median :3.090
Mean :3.135
3rd Qu.:3.490
Max. :5.170
bm <- brm(distance ~ social.influence * status, dat)
Compiling the C++ model
Error in compileCode(f, code, language = language, verbose = verbose) :
Compilation ERROR, function(s)/method(s) not created! In file included from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/BH/include/boost/smart_ptr/shared_ptr.hpp:28:0,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/BH/include/boost/shared_ptr.hpp:17,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat/fun/LDLT_factor.hpp:7,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat/err/check_ldlt_factor.hpp:6,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat.hpp:33,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/rev/mat.hpp:12,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math.hpp:4,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-libr
In addition: Warning message:
In system(cmd, intern = !verbose) :
Error in compileCode(f, code, language = language, verbose = verbose) :
Compilation ERROR, function(s)/method(s) not created! In file included from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/BH/include/boost/smart_ptr/shared_ptr.hpp:28:0,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/BH/include/boost/shared_ptr.hpp:17,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat/fun/LDLT_factor.hpp:7,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat/err/check_ldlt_factor.hpp:6,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat.hpp:33,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/rev/mat.hpp:12,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math.hpp:4,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-libr
Error in sink(type = "output") : invalid connection
What indicates that the issue is the ability to write/execute files in /tmp?
Far more likely is that the compilation is hitting the memory limit of 1 GB. I am going to test that theory myself.
Have you tried using one of the base projects that the Stan folks have created? Those are configured to use clang instead of gcc, which they say makes better use of the available memory.
I think the issue is executing the compiled C++ model because the same occurs on my local machine if I don't remount /tmp as exec. Besides, the data include few observations and I have 32G RAM, which apparently does not help.
But the error message is during compilation, not execution.
Given you have 32 GB locally and there is only 1 GB on rstudio.cloud, I still suspect that is the issue.
Would you be willing to DM me the CSV file you have in your example? I don't actually know how to use Stan, I just pretend really well by copying what others do. 
Here's a fully reproducible example. I do not really understand what is going on. It seems that some library is missing, but then why remounting /tmp should work? What I can say is that
- on my Xubuntu 18.04 machine, the C complier uses ~2G
- despite much more RAM, I get the same error if I don't remount the /tmp directory typing
sudo mount -o remount, exec /tmpin the terminal
Thank you for your help.
> library(brms)
Loading required package: Rcpp
Loading required package: ggplot2
Loading 'brms' package (version 2.7.0). Useful instructions
can be found by typing help('brms'). A more detailed introduction
to the package is available through vignette('brms_overview').
Run theme_set(theme_default()) to use the default bayesplot theme.
>
> x1 <- runif(300, 0, 100)
> x2 <- sample(0:1, 300, replace=T)
> y <- 2 * x1 + 3 * x2 + 0.5 * x1 * x2 + rnorm(300, 0, 10)
> df <- data.frame(x1, x2, y)
>
> bm <- brm(y ~ x1 * x2, df)
Compiling the C++ model
Error in compileCode(f, code, language = language, verbose = verbose) :
Compilation ERROR, function(s)/method(s) not created! In file included from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/BH/include/boost/smart_ptr/shared_ptr.hpp:28:0,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/BH/include/boost/shared_ptr.hpp:17,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat/fun/LDLT_factor.hpp:7,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat/err/check_ldlt_factor.hpp:6,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat.hpp:33,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/rev/mat.hpp:12,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math.hpp:4,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-libr
In addition: Warning message:
In system(cmd, intern = !verbose) :
Error in compileCode(f, code, language = language, verbose = verbose) :
Compilation ERROR, function(s)/method(s) not created! In file included from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/BH/include/boost/smart_ptr/shared_ptr.hpp:28:0,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/BH/include/boost/shared_ptr.hpp:17,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat/fun/LDLT_factor.hpp:7,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat/err/check_ldlt_factor.hpp:6,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/prim/mat.hpp:33,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math/rev/mat.hpp:12,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-library/3.5/StanHeaders/include/stan/math.hpp:4,
from /home/rstudio-user/R/x86_64-pc-linux-gnu-libr
Error in sink(type = "output") : invalid connection
I can confirm that it is running out of memory during compilation, and that using the base project the Stan folks put together that uses clang does not help.
I have no ideas about your situation with /tmp locally, but as you can see it is not a separate mountable device in rstudio.cloud:
rstudio-user@db406845f9a4:/cloud/project$ df -h /tmp
Filesystem Size Used Avail Use% Mounted on
none 125G 38G 87G 30% /
It is just part of the root filesystem.
This topic was automatically closed 21 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.