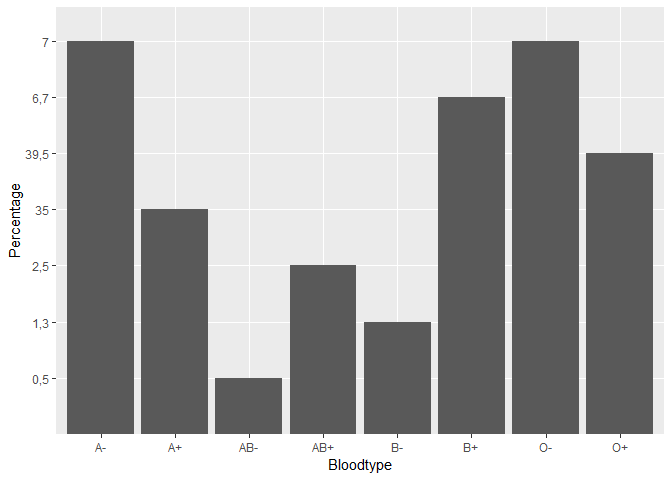
I have a plot of the distribution of blood types in the Netherlands, but my y-axis is a bit messy as the values of A+ and O- are much larger than the rest (see picture). How can I get this right?
This is my code in Rstudio:
ggplot(data = Blood_types_Sanquin, mapping = aes(x=Bloodtype, y=Percentage))+
geom_bar(stat = "Identity")![Bloodtype%20distribution|689x462]
(upload://umNMkgIBjVrJevj1bYUF03lXiiw.png)