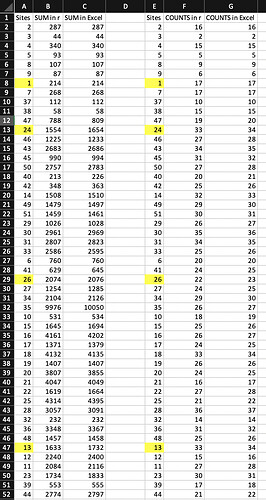
I've discovered an issue when calculating sum of rows and counts (">0") in my data frame. I'm trying to sum rows to have abundance and richness of a community data "spe" (51 columns(taxa) and 82 rows(sites)). I realize this when double checking the sums and counts in R and Excel. Why is this happening? This is the code I use in R:
For Sum:
apply(spe[,-1],1,sum)
For Counts:
apply(spe[,-1]>0,1,sum)
I've noticed the first rows goes well with the sums and counts, but then the results in R are different by subtracting numbers in sums and just 1 less in the counts issue, and after some rows, there are some good with the sums and counts but not all of them comparing to Excel. Why is that happening? Can someone help me please? Thanks in advance.
spe<-data.frame(read.table("~/Dropbox/Docs Redro/Paper Tesis MBC/MBC_SPP_ALL.csv", sep=",", header=TRUE, row.names=1))
head(spe)
#> Duge Allua1 Ande Anom Atop1 Atop2 Bleph1 Bleph2 Bleph3 Caillo2 Caillo3
#> 2 0 0 0 0 1 0 0 0 0 0 0
#> 3 0 0 0 0 0 0 0 0 0 0 0
#> 4 0 0 0 0 0 0 0 0 0 0 0
#> 5 0 0 0 0 0 0 0 0 0 0 0
#> 8 0 0 0 0 0 0 0 0 0 0 0
#> 9 0 0 0 0 0 0 0 0 0 0 0
#> Caillo1 Ceratoindet Cheli2 chiroindet Chiroinae Claudio Contu Corix2 Crambi
#> 2 2 0 1 0 0 0 0 0 0
#> 3 0 0 0 0 0 0 0 0 0
#> 4 3 0 2 0 0 0 0 0 0
#> 5 0 0 0 0 0 0 0 0 0
#> 8 6 0 0 0 0 0 0 0 0
#> 9 0 0 0 0 0 0 0 0 0
#> Curcu1 Curcu2 Diame1 Diame2 Dimec Dytis1 Ephyd2 Geran Helicho Hemer1 Hyale
#> 2 0 0 1 0 0 0 0 0 0 2 0
#> 3 0 0 0 0 0 0 0 0 0 0 0
#> 4 0 0 2 1 0 0 0 0 0 1 0
#> 5 0 0 0 0 0 0 0 0 0 0 0
#> 8 0 0 0 0 0 1 0 0 0 0 0
#> 9 0 0 1 0 0 0 0 0 0 0 0
#> HYDRA Hydrosc Leech1 Leech2 Leech3 Leech4 Leuco Limoniindet Lumbri Lymnae
#> 2 0 0 0 0 0 0 0 0 0 0
#> 3 0 0 0 0 0 0 0 0 0 0
#> 4 0 0 0 0 2 0 0 0 0 0
#> 5 0 0 0 0 0 0 0 0 0 0
#> 8 0 0 0 0 0 0 0 0 0 0
#> 9 0 0 0 0 0 0 0 0 0 0
#> Maya Molo1 Molo2 Morto Musci5 Musci6 Musci7 Musci4 Musci1 Musci2 Musci3 Naid1
#> 2 0 1 0 0 0 0 0 1 11 0 0 39
#> 3 0 0 0 0 0 0 0 0 0 0 0 0
#> 4 0 0 0 0 0 0 0 0 8 0 0 32
#> 5 0 0 0 0 0 0 0 1 0 0 0 0
#> 8 0 0 1 0 0 0 0 0 2 0 0 0
#> 9 0 0 0 0 0 0 0 0 0 0 0 0
#> Naid2 Naid3 Nectop Nema Neoel1 Neoel2 Neot Nepti Nona1 Ochro Ortho Ostra
#> 2 0 0 0 2 0 0 0 0 0 0 72 0
#> 3 0 0 0 0 0 0 0 0 0 0 0 0
#> 4 0 0 0 4 0 1 0 0 0 0 85 0
#> 5 0 0 0 0 0 0 0 0 0 0 7 0
#> 8 0 0 0 2 0 0 0 0 0 0 43 0
#> 9 0 0 0 0 0 0 0 0 0 0 2 0
#> Podo1 Podo2 Podo3 Podo4 Priono1 Priono2 Priono3 Schoe Simu1 Sphae Staphy1
#> 2 24 0 4 1 0 0 0 0 124 0 1
#> 3 16 0 28 0 0 0 0 0 0 0 0
#> 4 177 0 2 2 0 0 0 0 18 0 0
#> 5 13 0 71 1 0 0 0 0 0 0 0
#> 8 46 0 3 0 0 0 0 0 3 0 0
#> 9 49 0 20 14 0 0 0 0 0 0 1
#> Staphy2 Stilo1 Sync1 Tanyp Tipu Trichoco
#> 2 0 0 0 0 0 0
#> 3 0 0 0 0 0 0
#> 4 0 0 0 0 0 0
#> 5 0 0 0 0 0 0
#> 8 0 0 0 0 0 0
#> 9 0 0 0 0 0 0
# Spp Richness
apply(spe[,-1]>0,1,sum)
#> 2 3 4 5 8 9 1 7 37 38 47 24 46 43 45 50 40 42 14 49 51 29 30 31 33 6
#> 16 2 15 5 9 6 17 17 10 15 19 33 27 34 31 27 20 25 32 29 30 26 35 34 25 20
#> 41 26 27 34 35 10 15 16 17 18 19 20 21 22 25 28 32 36 48 13 12 11 23 39 44
#> 24 22 24 29 26 18 25 26 24 33 26 24 16 27 21 36 14 31 25 33 15 27 30 17 21
# Abundance
apply(spe[,-1],1,sum)
#> 2 3 4 5 8 9 1 7 37 38 47 24 46 43 45 50
#> 287 44 340 93 107 87 214 268 112 58 788 1554 1225 2683 990 2757
#> 40 42 14 49 51 29 30 31 33 6 41 26 27 34 35 10
#> 213 348 1508 1479 1459 1026 2961 2807 2586 760 629 2074 1254 2104 9976 531
#> 15 16 17 18 19 20 21 22 25 28 32 36 48 13 12 11
#> 1645 4161 1371 4132 1407 3807 4047 1619 4314 3057 232 3348 1457 1633 2240 2084
#> 23 39 44
#> 1734 553 2774
dput(head(spe))
#> structure(list(Duge = c(0L, 0L, 0L, 0L, 0L, 0L), Allua1 = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Ande = c(0L, 0L, 0L, 0L, 0L, 0L), Anom = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Atop1 = c(1L, 0L, 0L, 0L, 0L, 0L), Atop2 = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Bleph1 = c(0L, 0L, 0L, 0L, 0L, 0L), Bleph2 = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Bleph3 = c(0L, 0L, 0L, 0L, 0L, 0L), Caillo2 = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Caillo3 = c(0L, 0L, 0L, 0L, 0L, 0L), Caillo1 = c(2L,
#> 0L, 3L, 0L, 6L, 0L), Ceratoindet = c(0L, 0L, 0L, 0L, 0L, 0L),
#> Cheli2 = c(1L, 0L, 2L, 0L, 0L, 0L), chiroindet = c(0L, 0L,
#> 0L, 0L, 0L, 0L), Chiroinae = c(0L, 0L, 0L, 0L, 0L, 0L), Claudio = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Contu = c(0L, 0L, 0L, 0L, 0L, 0L), Corix2 = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Crambi = c(0L, 0L, 0L, 0L, 0L, 0L),
#> Curcu1 = c(0L, 0L, 0L, 0L, 0L, 0L), Curcu2 = c(0L, 0L, 0L,
#> 0L, 0L, 0L), Diame1 = c(1L, 0L, 2L, 0L, 0L, 1L), Diame2 = c(0L,
#> 0L, 1L, 0L, 0L, 0L), Dimec = c(0L, 0L, 0L, 0L, 0L, 0L), Dytis1 = c(0L,
#> 0L, 0L, 0L, 1L, 0L), Ephyd2 = c(0L, 0L, 0L, 0L, 0L, 0L),
#> Geran = c(0L, 0L, 0L, 0L, 0L, 0L), Helicho = c(0L, 0L, 0L,
#> 0L, 0L, 0L), Hemer1 = c(2L, 0L, 1L, 0L, 0L, 0L), Hyale = c(0L,
#> 0L, 0L, 0L, 0L, 0L), HYDRA = c(0L, 0L, 0L, 0L, 0L, 0L), Hydrosc = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Leech1 = c(0L, 0L, 0L, 0L, 0L, 0L),
#> Leech2 = c(0L, 0L, 0L, 0L, 0L, 0L), Leech3 = c(0L, 0L, 2L,
#> 0L, 0L, 0L), Leech4 = c(0L, 0L, 0L, 0L, 0L, 0L), Leuco = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Limoniindet = c(0L, 0L, 0L, 0L, 0L,
#> 0L), Lumbri = c(0L, 0L, 0L, 0L, 0L, 0L), Lymnae = c(0L, 0L,
#> 0L, 0L, 0L, 0L), Maya = c(0L, 0L, 0L, 0L, 0L, 0L), Molo1 = c(1L,
#> 0L, 0L, 0L, 0L, 0L), Molo2 = c(0L, 0L, 0L, 0L, 1L, 0L), Morto = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Musci5 = c(0L, 0L, 0L, 0L, 0L, 0L),
#> Musci6 = c(0L, 0L, 0L, 0L, 0L, 0L), Musci7 = c(0L, 0L, 0L,
#> 0L, 0L, 0L), Musci4 = c(1L, 0L, 0L, 1L, 0L, 0L), Musci1 = c(11L,
#> 0L, 8L, 0L, 2L, 0L), Musci2 = c(0L, 0L, 0L, 0L, 0L, 0L),
#> Musci3 = c(0L, 0L, 0L, 0L, 0L, 0L), Naid1 = c(39L, 0L, 32L,
#> 0L, 0L, 0L), Naid2 = c(0L, 0L, 0L, 0L, 0L, 0L), Naid3 = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Nectop = c(0L, 0L, 0L, 0L, 0L, 0L),
#> Nema = c(2L, 0L, 4L, 0L, 2L, 0L), Neoel1 = c(0L, 0L, 0L,
#> 0L, 0L, 0L), Neoel2 = c(0L, 0L, 1L, 0L, 0L, 0L), Neot = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Nepti = c(0L, 0L, 0L, 0L, 0L, 0L), Nona1 = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Ochro = c(0L, 0L, 0L, 0L, 0L, 0L), Ortho = c(72L,
#> 0L, 85L, 7L, 43L, 2L), Ostra = c(0L, 0L, 0L, 0L, 0L, 0L),
#> Podo1 = c(24L, 16L, 177L, 13L, 46L, 49L), Podo2 = c(0L, 0L,
#> 0L, 0L, 0L, 0L), Podo3 = c(4L, 28L, 2L, 71L, 3L, 20L), Podo4 = c(1L,
#> 0L, 2L, 1L, 0L, 14L), Priono1 = c(0L, 0L, 0L, 0L, 0L, 0L),
#> Priono2 = c(0L, 0L, 0L, 0L, 0L, 0L), Priono3 = c(0L, 0L,
#> 0L, 0L, 0L, 0L), Schoe = c(0L, 0L, 0L, 0L, 0L, 0L), Simu1 = c(124L,
#> 0L, 18L, 0L, 3L, 0L), Sphae = c(0L, 0L, 0L, 0L, 0L, 0L),
#> Staphy1 = c(1L, 0L, 0L, 0L, 0L, 1L), Staphy2 = c(0L, 0L,
#> 0L, 0L, 0L, 0L), Stilo1 = c(0L, 0L, 0L, 0L, 0L, 0L), Sync1 = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Tanyp = c(0L, 0L, 0L, 0L, 0L, 0L), Tipu = c(0L,
#> 0L, 0L, 0L, 0L, 0L), Trichoco = c(0L, 0L, 0L, 0L, 0L, 0L)), row.names = c(2L,
#> 3L, 4L, 5L, 8L, 9L), class = "data.frame")
Created on 2020-03-24 by the reprex package (v0.3.0)