nla.simp <- gbm.simplify(nla.tc5.lr005, n.drops = 5)
gbm.simplify - version 2.9
simplifying gbm.step model for BenPlankRat with 8 predictors and 38 observations
original deviance = 0.154(0.0381)
a fixed number of 5 drops will be tested
creating initial models...
Error in gbm.fit(x = x, y = y, offset = offset, distribution = distribution, :
The data set is too small or the subsampling rate is too large: nTrain * bag.fraction <= n.minobsinnode
Error: $ operator is invalid for atomic vectors
See the FAQ: How to do a minimal reproducible example reprex for beginners. It's very hard to debug code without representative data. All I can say is that nla.tc5.lr005, 38 observations may be too few (the gbm.step example in the docs uses 200, but fails at 38).
library(dismo)
#> Loading required package: raster
#> Loading required package: sp
data(Anguilla_train)
# reduce data set to speed things up a bit
Anguilla_train = Anguilla_train[1:200,]
# Anguilla_train = Anguilla_train[1:38,] # fails with same error in OP
angaus.tc5.lr01 <- gbm.step(data=Anguilla_train, gbm.x = 3:14, gbm.y = 2, family = "bernoulli",
tree.complexity = 5, learning.rate = 0.01, bag.fraction = 0.5)
#> Loading required namespace: gbm
#>
#>
#> GBM STEP - version 2.9
#>
#> Performing cross-validation optimisation of a boosted regression tree model
#> for Angaus and using a family of bernoulli
#> Using 200 observations and 12 predictors
#> creating 10 initial models of 50 trees
#>
#> folds are stratified by prevalence
#> total mean deviance = 1.0905
#> tolerance is fixed at 0.0011
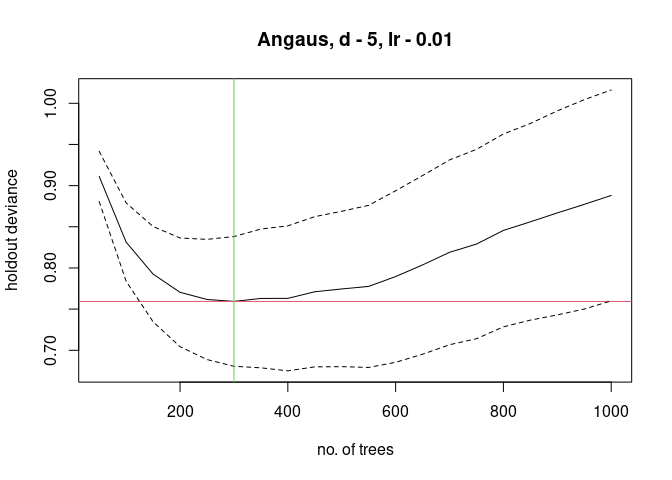
#> ntrees resid. dev.
#> 50 0.9112
#> now adding trees...
#> 100 0.8313
#> 150 0.7925
#> 200 0.7704
#> 250 0.7617
#> 300 0.7594
#> 350 0.763
#> 400 0.7631
#> 450 0.7711
#> 500 0.7745
#> 550 0.7776
#> 600 0.7896
#> 650 0.8037
#> 700 0.819
#> 750 0.829
#> 800 0.8456
#> 850 0.8561
#> 900 0.8669
#> 950 0.8773
#> 1000 0.8881
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> fitting final gbm model with a fixed number of 300 trees for Angaus
#>
#> mean total deviance = 1.09
#> mean residual deviance = 0.415
#>
#> estimated cv deviance = 0.759 ; se = 0.079
#>
#> training data correlation = 0.855
#> cv correlation = 0.545 ; se = 0.068
#>
#> training data AUC score = 0.985
#> cv AUC score = 0.867 ; se = 0.03
#>
#> elapsed time - 0.03 minutes
This topic was automatically closed 21 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.