Hi all!
I'm testing what effects chick feldging success (FledgeAD). Chicks who fledged successfully is '1' and those who died is '0'. Therefore I used glmer with binominal family. My fixed effect are mean temperature (meanT), individual number of chick (IndNum) and species (3 species). Random effect is season (3 breeding seasons).
For some reason the intercept for the fixed effects are not showing. Anyone know why and how I can get the intercept? The summary() output does not show the intercept of fixed effects
This is what I've tried thus far.
library(lme4)
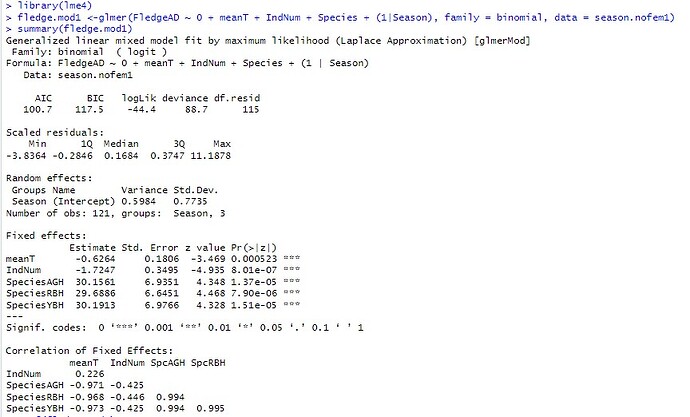
fledge.mod1 <-glmer(FledgeAD ~ 0 + meanT + IndNum + Species + (1|Season), family = binomial, data = season.nofem1)
summary(fledge.mod1)
Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod]
Family: binomial ( logit )
Formula: FledgeAD ~ 0 + meanT + IndNum + Species + (1 | Season)
Data: season.nofem1
AIC BIC logLik deviance df.resid
100.7 117.5 -44.4 88.7 115
Scaled residuals:
Min 1Q Median 3Q Max
-3.8364 -0.2846 0.1684 0.3747 11.1878
Random effects:
Groups Name Variance Std.Dev.
Season (Intercept) 0.5984 0.7735
Number of obs: 121, groups: Season, 3
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
meanT -0.6264 0.1806 -3.469 0.000523 ***
IndNum -1.7247 0.3495 -4.935 8.01e-07 ***
SpeciesAGH 30.1561 6.9351 4.348 1.37e-05 ***
SpeciesRBH 29.6886 6.6451 4.468 7.90e-06 ***
SpeciesYBH 30.1913 6.9766 4.328 1.51e-05 ***
Signif. codes: 0 ‘’ 0.001 ‘’ 0.01 ‘’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
meanT IndNum SpcAGH SpcRBH
IndNum 0.226
SpeciesAGH -0.971 -0.425
SpeciesRBH -0.968 -0.446 0.994
SpeciesYBH -0.973 -0.425 0.994 0.995