I recently saw a textbook about the tidymodels package. I want to use the colon dataset in the survival package for testing, but I don't know why I encountered a problem.
My code is as follows
library(tidymodels)
library(survival)
data(colon)
str(colon)
colon$sex <- ifelse(colon$sex==1,"male","female")
colon$obstruct <- ifelse(colon$obstruct ==1,"yes","no")
colon$perfor <- ifelse(colon$perfor ==1,"yes","no")
colon$adhere <- ifelse(colon$adhere ==1,"yes","no")
colon$status <- ifelse(colon$obstruct ==1,"death","alive")
colon$node4<- ifelse(colon$node4 ==1,"yes","no")
colon <- select(colon,id,age,rx,sex,age,obstruct,perfor,adhere,nodes,status)
colon <- na.omit(colon)
data_split<- initial_split(colon,
prop = 3/4,
strata = status)
train_data <- training(data_split)
test_data<- testing(data_split)
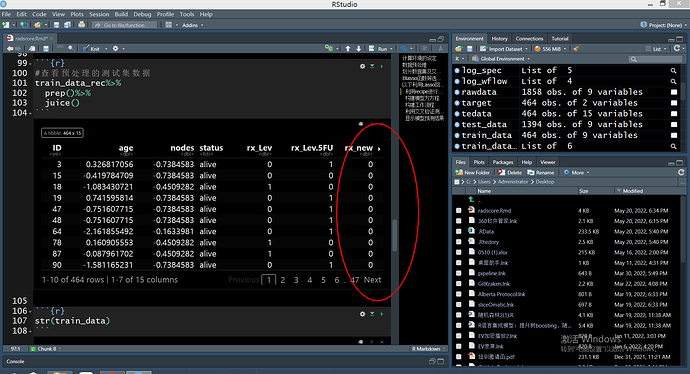
str(train_data)
train_rec <-
recipe(status ~., data = train_data) %>%
update_role(id, new_role = "ID")%>%
step_zv(all_numeric(),-all_outcomes()) %>%
step_normalize(all_numeric(),-all_outcomes())%>%
step_novel(all_nominal(),-all_outcomes()) %>%
step_dummy(all_nominal(),-all_outcomes())
summary(train_rec)
prepped_data <-
train_rec %>% # use the recipe object
prep() %>% # perform the recipe on training data
juice() # extract only the preprocessed dataframe
glimpse(prepped_data)
set.seed(100)
cv_folds <-
vfold_cv(train_data,
v = 5,
strata = status)
log_spec <- # your model specification
logistic_reg() %>% # model type
set_engine(engine = "glm") %>% # model engine
set_mode("classification") # model mode
log_wflow <- # new workflow object
workflow() %>% # use workflow function
add_recipe(train_rec) %>% # use the new recipe
add_model(log_spec) # add your model spec
log_res <-
log_wflow %>%
fit_resamples(
resamples = cv_folds,
metrics = metric_set(
precision, f_meas,
accuracy, kap,
roc_auc, sens, spec),
control = control_resamples(
save_pred = TRUE)
)
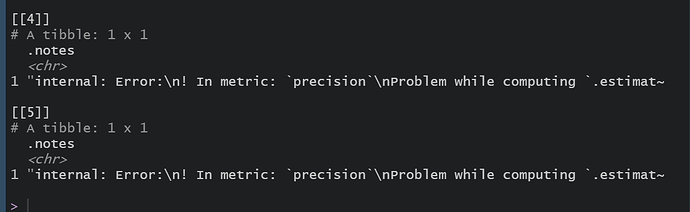
log_res$.notes
Is there a few things I don't quite understand, or is it a bug in this package?
- Why does my model not fit? I think it may be a problem with the recipe step.
- I already have the ID variable defined, why does step_normalize also normalize it?
- For the binary variable of gender, how does the sex_new that appears after dummy explain?