I know this isn't a coding question, so I apologize if it's too off topic. I'm confused because I don't understand why an ANOVA/linear model would give a different answer than VIF of a simple linear model with 2 predictor variables. For example, if I want to know if water temp. (a continuous covariate) is too closely related to a factor variable "season" to be put into the same model to predict some other variable (3rd variable not important here), what's the best way to tell? Obviously water temp. and season would seem too related, but choose any 2 variables! How does one test one continuous variable and a factor for similarities when it's not obvious? What is the best way to answer "are these two too closely related to be in the same model?"(?) Why isn't the VIF "test" telling me these are too related (VIF is less than 4!), like the linear model (at least in a different way)?
library(gratia)
library(performance)
df <- as.data.frame(rnorm(50, mean = 23.8, sd = 2.60))
df2 <- as.data.frame(rnorm(50, mean = 30.2, sd = 1.62))
names(df)[1] <- "temp"
names(df2)[1] <- "temp"
df3 <- rbind(df, df2)
df3$season <- rep(c("DRY", "WET"), each=50)
x <- rnorm(100, mean=0, sd=1)
df3$x <- x
full <- lm(x ~ temp + season, data=df3)
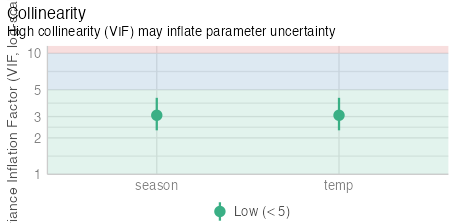
vif.cat.data <- check_collinearity(full)
vif.cat.data
# Check for Multicollinearity
Low Correlation
Term VIF VIF 95% CI Increased SE Tolerance Tolerance 95% CI
temp 3.04 [2.28, 4.23] 1.74 0.33 [0.24, 0.44]
season 3.04 [2.28, 4.23] 1.74 0.33 [0.24, 0.44]
# NOT too closely related!?
###############################################################################
# Now with ANOVA/linear model
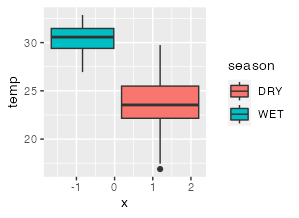
lm <- lm(temp ~ season, data=df3)
summary(lm)
Call:
lm(formula = temp ~ season, data = df3)
Residuals:
Min 1Q Median 3Q Max
-5.9874 -0.9893 -0.0597 1.1201 5.1884
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 24.8731 0.2729 91.14 <2e-16 ***
seasonWET 5.4516 0.3860 14.12 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.93 on 98 degrees of freedom
Multiple R-squared: 0.6706, Adjusted R-squared: 0.6672 # <--Definitely related??
F-statistic: 199.5 on 1 and 98 DF, p-value: < 2.2e-16