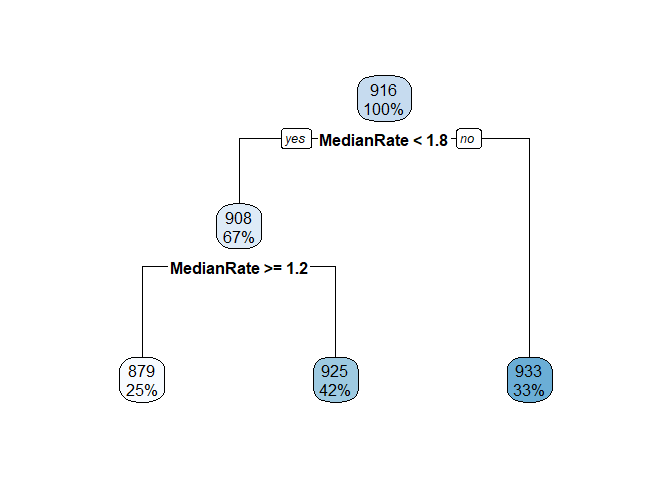
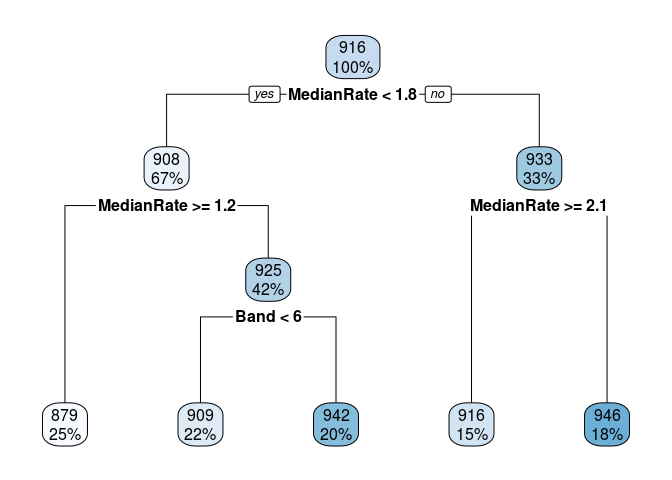
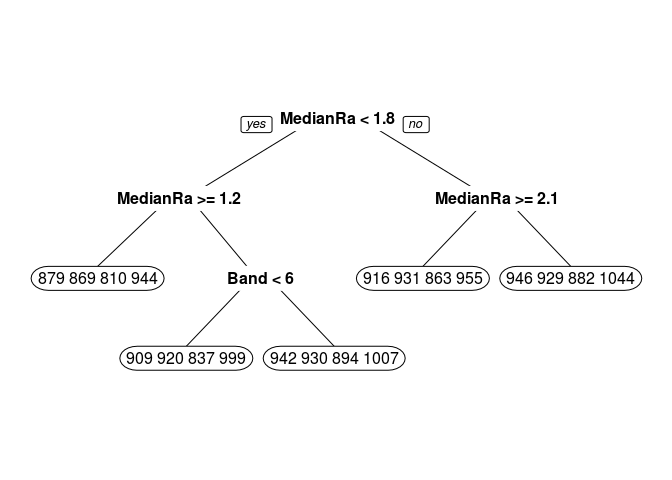
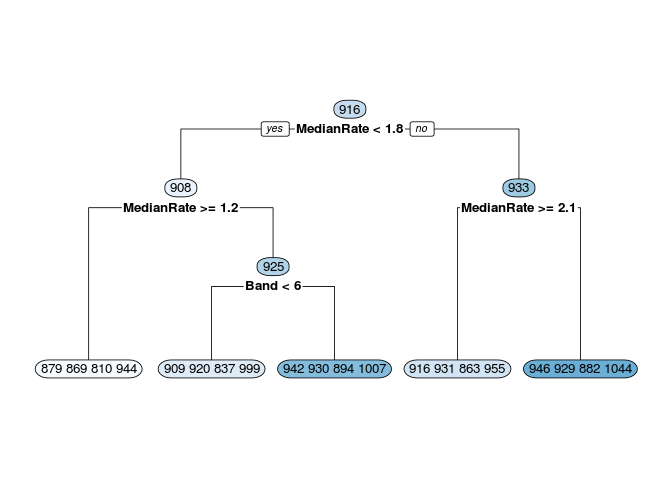
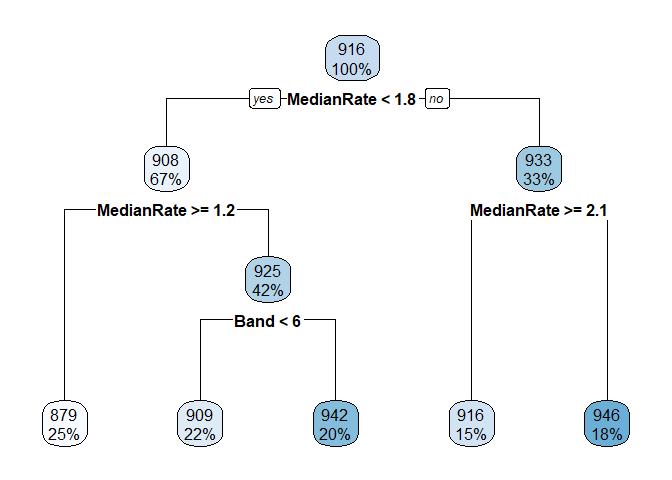
Below are some calculations on a data set I invented. Finding the mean, median, etc. of the leaf nodes is shown first. It is easy to match LeafStats$LeafNum to the rpart.plot by looking at the Mean values.
I started calculations for the other nodes and you can see that it is much uglier. From fit$splits I can get from the index column the exact value used to split each node. For example, the top node is split at MedianRate < 1.7635074. Competing splits that where not used are also shown in Splits, so you have to be careful. From the fit$frame element I can get the node numbers and the mean y values of each node.
My function GetStats() uses the NodeNum simply as a label in case you wanted to join the individual node data frames together.
If the tree has many nodes, these manual calculations would be very tedious. I do not see at the moment how to automate the calculations for the non-leaf nodes.
#Invent data
set.seed(1)
DF1 <- data.frame(Band = round(rnorm(30, 5, 0.7), 0),
MedianRate = runif(30, min = 0.25, max = 2.25),
Value = rnorm(30, 900, 50))
DF2 <- data.frame(Band = round(rnorm(30, 6, 0.7), 0),
MedianRate = runif(30, min = 0.5, max = 2.5),
Value = rnorm(30, 940, 50))
DF <- rbind(DF1, DF2)
library(rpart)
library(rpart.plot)
library(dplyr)
fit <- rpart(Value~Band+MedianRate, data=DF, method="anova")
rpart.plot(fit)
#Calculate statistics of the leaves
LeafStats <- DF %>% mutate(LeafNum = fit$where) %>%
group_by(LeafNum) %>%
summarise(Mean = mean(Value), Median = median(Value),
Min = min(Value), Max = max(Value), N = n())
LeafStats #See the print out of Frame below to understand LeafNum
#> # A tibble: 5 x 6
#> LeafNum Mean Median Min Max N
#> <int> <dbl> <dbl> <dbl> <dbl> <int>
#> 1 3 879. 869. 810. 944. 15
#> 2 5 909. 920. 837. 999. 13
#> 3 6 942. 930. 894. 1007. 12
#> 4 8 916. 931. 863. 955. 9
#> 5 9 946. 929. 882. 1044. 11
###Node Calculations
Splits <- fit$splits
Splits
#> count ncat improve index adj
#> MedianRate 60 -1 0.0578866182 1.7635074 0.0000000
#> Band 60 -1 0.0012291083 5.5000000 0.0000000
#> MedianRate 40 1 0.2155986209 1.1872099 0.0000000
#> Band 40 1 0.0002392061 5.5000000 0.0000000
#> Band 25 -1 0.1266509674 5.5000000 0.0000000
#> MedianRate 25 -1 0.1038963104 0.7519308 0.0000000
#> MedianRate 0 -1 0.7200000000 0.9077468 0.4166667
#> MedianRate 20 1 0.1026266061 2.0550743 0.0000000
#> Band 20 -1 0.0111632486 5.5000000 0.0000000
#> Band 0 1 0.6500000000 6.5000000 0.2222222
Frame <- fit$frame
Frame
#> var n wt dev yval complexity ncompete nsurrogate
#> 1 MedianRate 60 60 142627.676 916.1906 0.09688132 1 0
#> 2 MedianRate 40 40 89887.778 907.8959 0.09688132 1 0
#> 4 <leaf> 15 15 18247.778 879.4796 0.01000000 0 0
#> 5 Band 25 25 52260.318 924.9457 0.04640628 1 1
#> 10 <leaf> 13 13 30606.923 909.3128 0.01000000 0 0
#> 11 <leaf> 12 12 15034.576 941.8814 0.01000000 0 0
#> 3 MedianRate 20 20 44483.665 932.7800 0.03200787 1 1
#> 6 <leaf> 9 9 8638.785 916.0772 0.01000000 0 0
#> 7 <leaf> 11 11 31279.672 946.4460 0.01000000 0 0
GetStats <- function(x, NodeNum) {
summarise(x, Mean = mean(Value), Median = median(Value),
Min = min(Value), Max = max(Value),
N = n(), Perc = n()/nrow(DF)) %>%
mutate(Node = NodeNum)
}
Node2 <- DF %>% filter(MedianRate < Splits[1, "index"])
Node2Stats <- GetStats(Node2, NodeNum = 2)
Node2Stats
#> Mean Median Min Max N Perc Node
#> 1 907.8959 912.0706 809.7521 1007.152 40 0.6666667 2
Node3 <- DF %>% filter(MedianRate >= Splits[1, "index"])
Node3Stats <- GetStats(Node3, NodeNum = 3)
Node3Stats
#> Mean Median Min Max N Perc Node
#> 1 932.78 930.1466 862.8363 1044.358 20 0.3333333 3
Node4 <- Node2 %>% filter(MedianRate >= Splits[3, "index"])
Node4Stats <- GetStats(Node4, NodeNum = 4)
Node4Stats
#> Mean Median Min Max N Perc Node
#> 1 879.4796 869.3987 809.7521 944.0554 15 0.25 4
Node5 <- Node2 %>% filter(MedianRate < Splits[3, "index"])
Node5Stats <- GetStats(Node5, NodeNum = 5)
Node5Stats
#> Mean Median Min Max N Perc Node
#> 1 924.9457 923.7865 837.3183 1007.152 25 0.4166667 5
Created on 2019-10-11 by the reprex package (v0.3.0.9000)