Variant Visualization App
Authors: Shreya Sharma
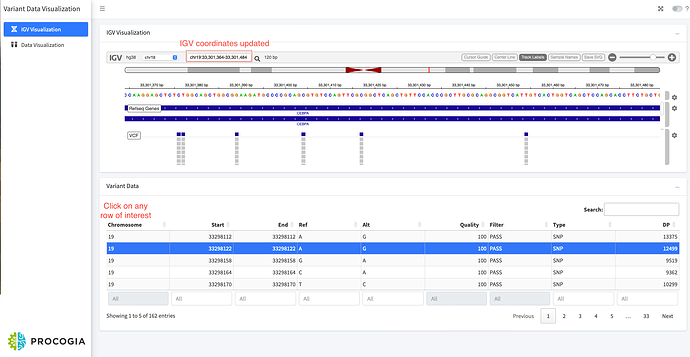
Abstract: The Variant Visualization application allows the user to interactively explore genomic variants in a VCF file. In the first tab, you'll find IGV for visualizing human genome variant data. The variant file is obtained from the sample data provided by the igvshiny R library and is from chromosome 19 CEBPA region. The user can click on a variant from the table, which will load the variant data and enable the user to view that variant location on IGV.
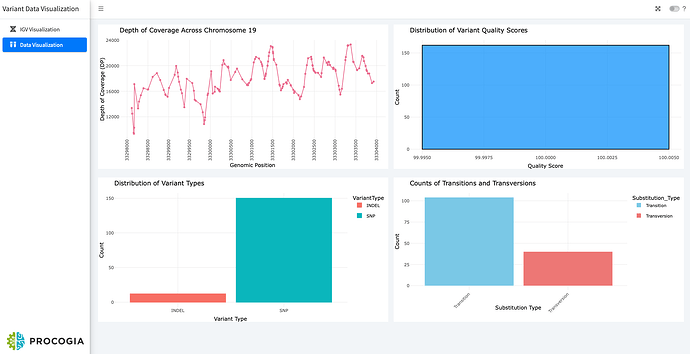
The second tab features various plots to explore the data: a scatter plot of coverage depth across the chromosome, a histogram of variant quality scores, and bar plots showing SNPs vs. INDELS and transitions vs. transversions.
Dive in to explore and visualize the variant data seamlessly!
Full Description:
Background
- VCF File
- A VCF (Variant Call Format) file is a standard text format used to store information about genetic variants, such as single nucleotide polymorphisms (SNPs) and insertions/deletions (INDELs) within a genome.
- It includes details like the variant’s position, type, and quality.
- The variant can be classified as a SNP or INDEL. SNPs (Single Nucleotide Polymorphisms) are variations at a single nucleotide position in the DNA sequence, while INDELs (Insertions/Deletions) refer to the addition or removal of small segments of DNA.
- Both types of variants can influence gene function and are crucial for studying genetic diversity and disease.
- Integrative Genomics Viewer (IGV)
- IGV is a powerful tool for visualizing genomic data.
- It provides a graphical interface to examine the genomic context of variants, such as their exact locations, the surrounding sequence, and coverage depth.
- This helps in understanding how variants might impact gene function or contribute to diseases.
Variant Visualization App Details
- This application uses the variant file obtained from the sample data provided by the
igvshinyR library and is from chromosome 19 CEBPA region. - The first tab of the application loads the variant data in IGV. User can select the row corresponding to the variant of interest in the variant table to visualize the variant in IGV. Once the user selects a row, the IGV coordinates are updated and the user can visualize the variant in that region.
- In the second tab, the app explores the variant data by constructing the following plots:
- Scatter plot displaying depth of coverage across the entire chromosome.
- Histogram indicating the distribution of variant quality scores.
- Bar plot indicating the count of SNPs and INDELS in the VCF file.
- Bar plot indicating the transitions and transversions in the data.
Conclusion
This app enables the user to interactively visualize the variant data for chromosome 19 CEBPA region.
Shiny app: Posit Connect Cloud
Repo: GitHub - shreya-sharma20/variantVisualizationApp: R Shiny app to visualize VCF file in IGV
Thumbnail:
Full image: