My recent research needs to compare different machine learning survival analysis modeling methods. I used the tidymodels package on classification problems and felt very good. I found relevant tutorials on Google, and the website is as follows: Posts /2021/11/survival-analysis-parsnip-adjacent/.
rm(list = ls())
options(stringsAsFactors = T)
#> Warning in options(stringsAsFactors = T): 'options(stringsAsFactors = TRUE)' is
#> deprecated and will be disabled
Sys.setenv(LANGUAGE = "en")
library(tidyverse)
#> Warning: package 'tidyverse' was built under R version 4.0.5
#> Warning: package 'ggplot2' was built under R version 4.0.5
#> Warning: package 'tidyr' was built under R version 4.0.5
#> Warning: package 'readr' was built under R version 4.0.5
#> Warning: package 'dplyr' was built under R version 4.0.5
#> Warning: package 'forcats' was built under R version 4.0.5
library(tidymodels)
#> Warning: package 'tidymodels' was built under R version 4.0.5
#> Registered S3 method overwritten by 'tune':
#> method from
#> required_pkgs.model_spec parsnip
#> Warning: package 'broom' was built under R version 4.0.5
#> Warning: package 'dials' was built under R version 4.0.5
#> Warning: package 'infer' was built under R version 4.0.5
#> Warning: package 'modeldata' was built under R version 4.0.5
#> Warning: package 'recipes' was built under R version 4.0.5
#> Warning: package 'tune' was built under R version 4.0.5
#> Warning: package 'workflows' was built under R version 4.0.5
#> Warning: package 'workflowsets' was built under R version 4.0.5
#> Warning: package 'yardstick' was built under R version 4.0.5
library(censored)
#> Loading required package: survival
#> Warning: package 'survival' was built under R version 4.0.5
library(survival)
# 载入数据集
bladder_train <- bladder[-c(1:3),]
bladder_test <- bladder[1:3,]
cox_spec <- proportional_hazards(penalty = 0.123) %>%
set_engine("glmnet")
f_fit <- fit(cox_spec,
Surv(stop, event) ~ rx + size + number + strata(enum),
data = bladder_train)
#> Error: 'stratifySurv' is not an exported object from 'namespace:glmnet'
f_pred <- predict(f_fit, new_data = bladder_test,
type = "survival", time = seq(0, 20, 1))
#> Error in predict(f_fit, new_data = bladder_test, type = "survival", time = seq(0, : object 'f_fit' not found
f_pred <- f_pred %>%
mutate(id = factor(1:3)) %>%
unnest(cols = .pred)
#> Error in mutate(., id = factor(1:3)): object 'f_pred' not found
f_pred %>%
ggplot(aes(x = .time, y = .pred_survival, col = id)) +
geom_step()
#> Error in ggplot(., aes(x = .time, y = .pred_survival, col = id)): object 'f_pred' not found
Hi @zhangkai ,
I am unable to reproduce your issue. What version of censored and parsnip are you using? Can you try updating the censored package and try again?
library(tidyverse)
library(tidymodels)
library(censored)
#> Loading required package: survival
library(survival)
bladder_train <- bladder[-c(1:3),]
bladder_test <- bladder[1:3,]
cox_spec <-
proportional_hazards(penalty = 0.123) %>%
set_engine("glmnet")
f_fit <- fit(cox_spec,
Surv(stop, event) ~ rx + size + number + strata(enum),
data = bladder_train)
f_pred <- predict(f_fit, new_data = bladder_test,
type = "survival", time = seq(0, 20, 1))
f_pred <- f_pred %>%
mutate(id = factor(1:3)) %>%
unnest(cols = .pred)
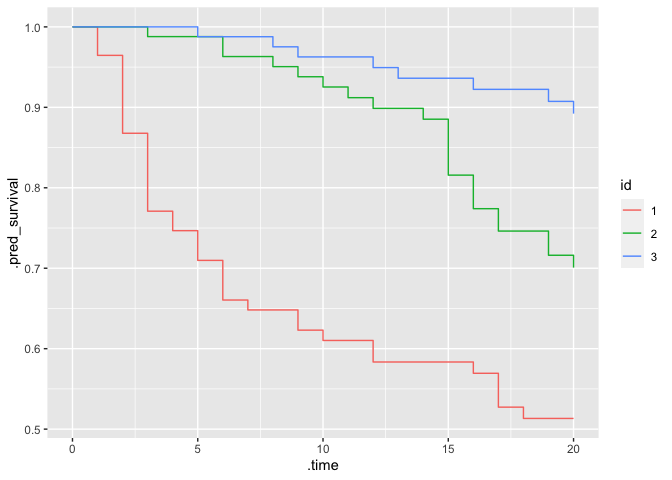
f_pred %>%
ggplot(aes(x = .time, y = .pred_survival, col = id)) +
geom_step()
Session info
sessioninfo::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.0.5 (2021-03-31)
#> os macOS Big Sur 10.16
#> system x86_64, darwin17.0
#> ui X11
#> language (EN)
#> collate en_CA.UTF-8
#> ctype en_CA.UTF-8
#> tz America/Toronto
#> date 2022-07-29
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date lib
#> assertthat 0.2.1 2019-03-21 [1]
#> backports 1.4.1 2021-12-13 [1]
#> broom * 0.7.12 2022-01-28 [1]
#> cellranger 1.1.0 2016-07-27 [1]
#> censored * 0.1.0 2022-06-22 [1]
#> class 7.3-18 2021-01-24 [2]
#> cli 3.3.0 2022-04-25 [1]
#> codetools 0.2-18 2020-11-04 [2]
#> colorspace 2.0-3 2022-02-21 [1]
#> crayon 1.5.1 2022-03-26 [1]
#> curl 4.3.2 2021-06-23 [1]
#> DBI 1.1.2 2021-12-20 [1]
#> dbplyr 2.1.1 2021-04-06 [1]
#> dials * 1.0.0 2022-06-14 [1]
#> DiceDesign 1.9 2021-02-13 [1]
#> digest 0.6.29 2021-12-01 [1]
#> dplyr * 1.0.99.9000 2022-07-29 [1]
#> ellipsis 0.3.2 2021-04-29 [1]
#> evaluate 0.15 2022-02-18 [1]
#> fansi 1.0.3 2022-03-24 [1]
#> farver 2.1.1 2022-07-06 [1]
#> fastmap 1.1.0.9000 2021-12-10 [1]
#> forcats * 0.5.0 2020-03-01 [1]
#> foreach 1.5.2 2022-02-02 [1]
#> fs 1.5.2.9000 2022-04-12 [1]
#> furrr 0.3.0 2022-05-04 [1]
#> future 1.26.1 2022-05-27 [1]
#> future.apply 1.9.0 2022-04-25 [1]
#> generics 0.1.3 2022-07-05 [1]
#> ggplot2 * 3.3.6 2022-05-03 [1]
#> glmnet 4.1-4 2022-04-15 [1]
#> globals 0.15.1-9006 2022-07-29 [1]
#> glue 1.6.2 2022-02-24 [1]
#> gower 1.0.0 2022-02-03 [1]
#> GPfit 1.0-8 2019-02-08 [1]
#> gtable 0.3.0 2019-03-25 [1]
#> hardhat 1.2.0.9000 2022-07-07 [1]
#> haven 2.3.1 2020-06-01 [1]
#> highr 0.9 2021-04-16 [1]
#> hms 1.1.0 2021-05-17 [1]
#> htmltools 0.5.2 2021-08-25 [1]
#> httr 1.4.3 2022-05-04 [1]
#> infer * 1.0.0 2021-08-13 [1]
#> ipred 0.9-13 2022-06-02 [1]
#> iterators 1.0.14 2022-02-05 [1]
#> jsonlite 1.8.0 2022-02-22 [1]
#> knitr 1.39 2022-04-26 [1]
#> labeling 0.4.2 2020-10-20 [1]
#> lattice 0.20-41 2020-04-02 [2]
#> lava 1.6.10 2021-09-02 [1]
#> lhs 1.1.5 2022-03-22 [1]
#> lifecycle 1.0.1 2021-09-24 [1]
#> listenv 0.8.0 2019-12-05 [1]
#> lubridate 1.8.0 2021-10-07 [1]
#> magrittr 2.0.3 2022-03-30 [1]
#> MASS 7.3-53.1 2021-02-12 [2]
#> Matrix 1.3-2 2021-01-06 [2]
#> mime 0.12 2021-09-28 [1]
#> modeldata * 0.1.1 2021-07-14 [1]
#> modelr 0.1.8 2020-05-19 [1]
#> munsell 0.5.0 2018-06-12 [1]
#> nnet 7.3-15 2021-01-24 [2]
#> parallelly 1.32.0 2022-06-07 [1]
#> parsnip * 1.0.0.9000 2022-07-29 [1]
#> pillar 1.8.0 2022-07-18 [1]
#> pkgconfig 2.0.3 2019-09-22 [1]
#> prodlim 2019.11.13 2019-11-17 [1]
#> purrr * 0.3.4.9000 2022-03-21 [1]
#> R6 2.5.1 2021-08-19 [1]
#> Rcpp 1.0.8.3 2022-03-17 [1]
#> readr * 2.1.2 2022-01-30 [1]
#> readxl 1.3.1 2019-03-13 [1]
#> recipes * 1.0.0.9000 2022-07-07 [1]
#> reprex 1.0.0 2021-01-27 [1]
#> rlang 1.0.4.9000 2022-07-29 [1]
#> rmarkdown 2.11.22 2022-02-22 [1]
#> rpart 4.1-15 2019-04-12 [1]
#> rsample * 1.0.0.9000 2022-07-07 [1]
#> rstudioapi 0.13 2020-11-12 [1]
#> rvest 1.0.1 2021-07-26 [1]
#> scales * 1.2.0 2022-04-13 [1]
#> sessioninfo 1.1.1 2018-11-05 [1]
#> shape 1.4.6 2021-05-19 [1]
#> stringi 1.7.6 2021-11-29 [1]
#> stringr * 1.4.0 2019-02-10 [1]
#> styler 1.3.2 2020-02-23 [1]
#> survival * 3.3-1 2022-03-03 [1]
#> tibble * 3.1.8 2022-07-22 [1]
#> tidymodels * 0.2.0.9000 2022-05-05 [1]
#> tidyr * 1.2.0.9000 2022-07-05 [1]
#> tidyselect 1.1.2.9000 2022-07-29 [1]
#> tidyverse * 1.3.0 2019-11-21 [1]
#> timeDate 3043.102 2018-02-21 [1]
#> tune * 0.2.0.9002 2022-07-07 [1]
#> tzdb 0.2.0 2021-10-27 [1]
#> utf8 1.2.2 2021-07-24 [1]
#> vctrs 0.4.1.9000 2022-07-29 [1]
#> withr 2.5.0 2022-03-03 [1]
#> workflows * 1.0.0.9000 2022-07-07 [1]
#> workflowsets * 0.2.1.9000 2022-07-07 [1]
#> xfun 0.30 2022-03-02 [1]
#> xml2 1.3.3 2021-11-30 [1]
#> yaml 2.3.5 2022-02-21 [1]
#> yardstick * 1.0.0.9000 2022-07-07 [1]
#> source
#> CRAN (R 4.0.0)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.0)
#> RSPM (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.2)
#> RSPM (R 4.0.5)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.2)
#> Github (tidyverse/dplyr@864fdc3)
#> CRAN (R 4.0.3)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> RSPM (R 4.0.5)
#> Github (r-lib/fastmap@d2722e7)
#> CRAN (R 4.0.0)
#> CRAN (R 4.0.5)
#> Github (r-lib/fs@e7d98c4)
#> CRAN (R 4.0.5)
#> RSPM (R 4.0.5)
#> CRAN (R 4.0.5)
#> RSPM (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> Github (HenrikBengtsson/globals@2629913)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.0)
#> CRAN (R 4.0.0)
#> Github (tidymodels/hardhat@1e43f0e)
#> CRAN (R 4.0.0)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.3)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.2)
#> RSPM (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.0)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.0)
#> CRAN (R 4.0.0)
#> CRAN (R 4.0.5)
#> RSPM (R 4.0.5)
#> Github (tidymodels/parsnip@f8505bd)
#> RSPM (R 4.0.5)
#> CRAN (R 4.0.0)
#> CRAN (R 4.0.0)
#> Github (tidyverse/purrr@d4b7ad1)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.0)
#> Github (tidymodels/recipes@372434c)
#> CRAN (R 4.0.2)
#> Github (r-lib/rlang@ea70713)
#> Github (rstudio/rmarkdown@72062ed)
#> CRAN (R 4.0.2)
#> Github (tidymodels/rsample@86f56df)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.0)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.0)
#> RSPM (R 4.0.5)
#> RSPM (R 4.0.5)
#> Github (tidymodels/tidymodels@5827b6a)
#> Github (tidyverse/tidyr@944f5ad)
#> Github (r-lib/tidyselect@190f80d)
#> CRAN (R 4.0.0)
#> CRAN (R 4.0.0)
#> Github (tidymodels/tune@d7d0edb)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.2)
#> Github (r-lib/vctrs@fe37f93)
#> CRAN (R 4.0.5)
#> Github (tidymodels/workflows@822f144)
#> Github (tidymodels/workflowsets@c58315f)
#> CRAN (R 4.0.5)
#> CRAN (R 4.0.2)
#> CRAN (R 4.0.5)
#> Github (tidymodels/yardstick@90ab794)
#>
#> [1] /Users/matt/Library/R/4.0/library
#> [2] /Library/Frameworks/R.framework/Versions/4.0/Resources/library
zhangkai:
strata(enum)
I updated the two packages you mentioned, but there is still a problem. The following is the version of the local program. I don't know if the problem lies in the glmnet package.
R version 4.0.3 (2020-10-10)
Matrix products: default
locale:
attached base packages:
other attached packages:
loaded via a namespace (and not attached):
Max
July 30, 2022, 2:07pm
4
I'd update the glmnet package. It is currently at 4.1-4
Thank you very much for your answer, it was indeed a problem with the glmnet package, the above problem was solved after the update
system
August 7, 2022, 12:53am
6
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.