Hello Community,
I have a dataframe with 10 columns, the first column is the treatment (4 treatments: C, H1, H2, H3), the rest of the columns (2:10) are variables (all numerical). I want to loop through each variable column and run an anova by the treatment types. So in the end I know "For variable X the variance is about the same between treatments, but for variable Y the variance is different, etc." I have no idea how to do this, I am new to this, so please be kind.
Thanks, Hannah
Without a reprex an example solution cannot be provided. This recent post shows the use of purrr::map to apply the same function to multiple objects.
Thank you. I don't have any code written as I cannot even figure out a way into the problem. I will review your recommendation.
Exemplary data suffices. It doesn't have to be actual data or a complete set, just representative.
Is this what you mean? (and thank you from the bottom of my heart)
| HarvTMT | FrondCount | Wt | MaxHt | MeanHt | SDHt | Girth | SA | FD | MeanOpen | SDOpen | OpenCount |
|---|---|---|---|---|---|---|---|---|---|---|---|
| H34 | 43 | 390 | 128 | 25.55813953 | 27.41159151 | 4 | 270 | 1.387 | 0.934733333 | 1.191087447 | 30 |
| H34 | 74 | 428 | 113 | 26.63513514 | 23.10522346 | 15 | 375 | 1.329 | 0.637329787 | 0.976997368 | 94 |
| H34 | 133 | 1304 | 87 | 27.09774436 | 20.1512706 | 35 | 606 | 1.491 | 0.651531646 | 1.059682486 | 79 |
| H34 | 68 | 4816 | 164 | 55.32352941 | 43.61002253 | NA | 732 | 1.538 | 1.067990291 | 2.2876416 | 103 |
| H34 | 66 | 486 | 71 | 29.95454545 | 16.34007044 | 19 | 100 | 1.378 | 0.559829787 | 0.596602958 | 94 |
| H34 | 29 | 304 | 95 | 45.96551724 | 18.50961673 | 15 | 91 | 1.338 | 0.890075758 | 1.331683727 | 66 |
| H34 | 36 | 1580 | 162 | 51.94444444 | 46.90412376 | 31 | 265 | 1.481 | NA | NA | NA |
| H34 | 22 | 2180 | 187 | 68.81818182 | 47.52707135 | 46 | 257 | 1.498 | NA | NA | NA |
| H1 | 31 | 172 | 57 | 28.48387097 | 18.31733417 | 15 | NA | NA | 0.592324324 | 1.059395662 | 37 |
| H1 | 13 | 200 | 105 | 40.46153846 | 31.36509574 | 10 | 44 | 1.285 | 0.856378049 | 1.38034015 | 82 |
| H1 | 21 | 39 | 24 | 28.61904762 | 10.81423224 | 36 | 41 | 1.22 | 0.611555556 | 0.96637902 | 27 |
| H1 | 38 | 606 | 94 | 25.39473684 | 16.9076018 | 20 | 112 | 1.459 | 0.601591549 | 1.025333529 | 71 |
| C | 140 | 2652 | 232 | 35.73571429 | 43.48250466 | 36 | 602 | 1.497 | 0.743346154 | 0.796051369 | 104 |
| C | 152 | 1334 | 111 | 26.60526316 | 24.97154882 | 31 | 361 | 1.438 | 3.665395349 | 16.95491841 | 86 |
| C | 51 | 372 | 87 | 20.39215686 | 18.36200254 | 18 | 85 | 1.345 | 0.59872043 | 0.779877783 | 93 |
| C | 240 | 14210 | NA | 43.7625 | 33.58218935 | NA | 972 | 1.513 | 0.344058824 | 0.419396714 | 187 |
| H2 | 65 | 838 | 68 | 18.06153846 | 19.94749117 | 30 | 440 | 1.43 | 0.583672897 | 0.948604633 | 107 |
| H2 | 55 | 2018 | 107 | 33.87272727 | 29.61797161 | 39 | 328 | 1.53 | 0.57144186 | 0.893088134 | 86 |
| H2 | 58 | 250 | 83 | 30.18965517 | 18.42810296 | 14 | 189 | 1.261 | 0.586148148 | 1.144894174 | 27 |
1 Like
# LIBRARIES
suppressPackageStartupMessages({library(dplyr)
library(ggplot2)
library(purrr)
library(readr)})
# DATA
read_csv("/home/roc/Desktop/grist.csv") -> dat
#> Parsed with column specification:
#> cols(
#> HarvTMT = col_character(),
#> FrondCount = col_double(),
#> Wt = col_double(),
#> MaxHt = col_double(),
#> MeanHt = col_double(),
#> SDHt = col_double(),
#> Girth = col_double(),
#> SA = col_double(),
#> FD = col_double(),
#> MeanOpen = col_double(),
#> SDOpen = col_double(),
#> OpenCount = col_double()
#> )
#> Warning: 1 parsing failure.
#> row col expected actual file
#> 19 -- 12 columns 7 columns '/home/roc/Desktop/grist.csv'
# PREPROCCESSING
# use rm_na(dat) -> dat, if desired
# CONSTANTS
the_vars <- colnames(dat)[2:length(dat)]
# FUNCTIONS
# eyeball reasonableness of distribution assumption
# use interactively
mk_plot <- function(x) {
p <- ggplot(dat)
p + geom_density(aes(x, fill = HarvTMT),
position = "identity", alpha = 0.5) +
labs(x = enquo(x), y = 'Density') +
scale_fill_discrete(name = 'Treatment') +
theme_minimal()
}
# map over the_vars
mk_aov <- function() {
dat[the_vars] %>% map(~ aov(. ~ dat$HarvTMT))
}
# remove NAs
rm_na <- function(x) {
x <- x[complete.cases(x),]
}
# test balance
test_bal <- function(x) {
!is.list(replications(~ x - as.factor(HarvTMT), dat))
}
# MAIN
# Example interactive plot
mk_plot(dat$FrondCount)
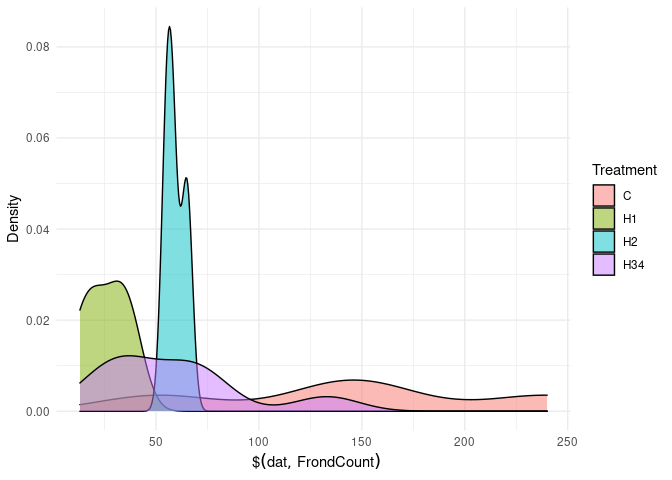
# map over the_vars to stdout
mk_aov()
#> $FrondCount
#> Call:
#> aov(formula = . ~ dat$HarvTMT)
#>
#> Terms:
#> dat$HarvTMT Residuals
#> Sum of Squares 32114.64 27233.04
#> Deg. of Freedom 3 15
#>
#> Residual standard error: 42.60911
#> Estimated effects may be unbalanced
#>
#> $Wt
#> Call:
#> aov(formula = . ~ dat$HarvTMT)
#>
#> Terms:
#> dat$HarvTMT Residuals
#> Sum of Squares 44679318 142793779
#> Deg. of Freedom 3 15
#>
#> Residual standard error: 3085.383
#> Estimated effects may be unbalanced
#>
#> $MaxHt
#> Call:
#> aov(formula = . ~ dat$HarvTMT)
#>
#> Terms:
#> dat$HarvTMT Residuals
#> Sum of Squares 13476.07 29081.54
#> Deg. of Freedom 3 14
#>
#> Residual standard error: 45.57689
#> Estimated effects may be unbalanced
#> 1 observation deleted due to missingness
#>
#> $MeanHt
#> Call:
#> aov(formula = . ~ dat$HarvTMT)
#>
#> Terms:
#> dat$HarvTMT Residuals
#> Sum of Squares 621.3159 2467.3116
#> Deg. of Freedom 3 15
#>
#> Residual standard error: 12.82527
#> Estimated effects may be unbalanced
#>
#> $SDHt
#> Call:
#> aov(formula = . ~ dat$HarvTMT)
#>
#> Terms:
#> dat$HarvTMT Residuals
#> Sum of Squares 423.081 1899.687
#> Deg. of Freedom 3 15
#>
#> Residual standard error: 11.2537
#> Estimated effects may be unbalanced
#>
#> $Girth
#> Call:
#> aov(formula = . ~ dat$HarvTMT)
#>
#> Terms:
#> dat$HarvTMT Residuals
#> Sum of Squares 152.0847 2113.7976
#> Deg. of Freedom 3 13
#>
#> Residual standard error: 12.75146
#> Estimated effects may be unbalanced
#> 2 observations deleted due to missingness
#>
#> $SA
#> Call:
#> aov(formula = . ~ dat$HarvTMT)
#>
#> Terms:
#> dat$HarvTMT Residuals
#> Sum of Squares 338043.8 796718.7
#> Deg. of Freedom 3 13
#>
#> Residual standard error: 247.5602
#> Estimated effects may be unbalanced
#> 2 observations deleted due to missingness
#>
#> $FD
#> Call:
#> aov(formula = . ~ dat$HarvTMT)
#>
#> Terms:
#> dat$HarvTMT Residuals
#> Sum of Squares 0.04038058 0.09870342
#> Deg. of Freedom 3 13
#>
#> Residual standard error: 0.08713536
#> Estimated effects may be unbalanced
#> 2 observations deleted due to missingness
#>
#> $MeanOpen
#> Call:
#> aov(formula = . ~ dat$HarvTMT)
#>
#> Terms:
#> dat$HarvTMT Residuals
#> Sum of Squares 1.252191 7.557395
#> Deg. of Freedom 3 12
#>
#> Residual standard error: 0.7935886
#> Estimated effects may be unbalanced
#> 3 observations deleted due to missingness
#>
#> $SDOpen
#> Call:
#> aov(formula = . ~ dat$HarvTMT)
#>
#> Terms:
#> dat$HarvTMT Residuals
#> Sum of Squares 38.92192 200.83808
#> Deg. of Freedom 3 12
#>
#> Residual standard error: 4.091028
#> Estimated effects may be unbalanced
#> 3 observations deleted due to missingness
#>
#> $OpenCount
#> Call:
#> aov(formula = . ~ dat$HarvTMT)
#>
#> Terms:
#> dat$HarvTMT Residuals
#> Sum of Squares 8582.167 12501.583
#> Deg. of Freedom 3 12
#>
#> Residual standard error: 32.27691
#> Estimated effects may be unbalanced
#> 3 observations deleted due to missingness
Created on 2020-09-15 by the reprex package (v0.3.0)
Wow, that's incredible. Thank you.
1 Like
This topic was automatically closed 21 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.