Dear all,
I am trying to implement the work done by Suyu Liu, “A Bayesian Phase I/II Trial Design for Immunotherapy”, using R, since the code attached with that work takes a lot of time (more than 20 hours and the code not complete, I do not know how it produced the tables and the figures).
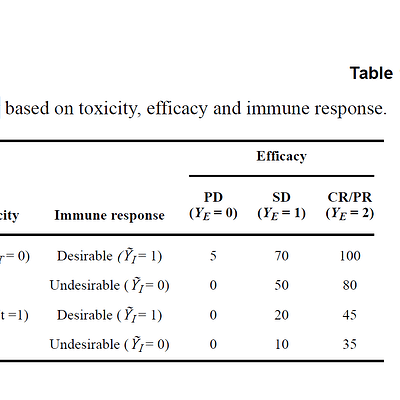
So that I tried to use trialr package trying to get similar or approximate results using the utility functions for sensitivity analysis in table 1 (picture below), but this package allowed me to use just two outcomes ( toxicity, efficacy ) and the work of Liu used three outcomes (immune response, toxicity, and efficacy). I want to see if I used the correct utility (table 1 below) and to see how to add a third outcome ( immune response ) to the model? (Question 1)
I attached to you the code and the output for 50 iterations and 60 patients from the work of Liu and Yuan ( I cannot do more, it took around 12 hours), if you know how he produced the results, (Question 2) I hope you can feed me back.
My goal is : to use their idea to select the best dose in the first stage and to continue in a second stage with only 2 arms clinical trial and the best dose. (may there is another way to do that?)
### My code trying to get similar results using trialr package ###
rm(list = ls())
library(trialr)
## Utility from table 1 and Liu and Yuan work.
Uti <- array(0,c(2,3,2)) # order: tox, eff, immuno
Uti[,,1] <- matrix(c(0,0,50,10,80,35),nrow=2)
Uti[,,2] <- matrix(c(5,0,70,20,100,45),nrow=2)
N.max= 60 # patients
outcomes <- '1NNN 2NNT 3NNT 4NNN 5NTN'
doses = c(.1,.3,.5,.7,.9)
fit <- stan_efftox(outcomes,
real_doses =doses,
efficacy_hurdle = 0.5, toxicity_hurdle = 0.3,
p_e = 0.1, p_t = 0.1,
eff0 = 0.5, tox1 = 0.65,
eff_star = 0.7, tox_star = 0.25,
alpha_mean = -7.9593, alpha_sd = 3.5487,
beta_mean = 1.5482, beta_sd = 3.5018,
gamma_mean = 0.7367, gamma_sd = 2.5423,
zeta_mean = 3.4181, zeta_sd = 2.4406,
eta_mean = 0, eta_sd = 0.2,
psi_mean = 0, psi_sd = 1,
seed = 123)
ndoses <- length(fit$prob_tox)
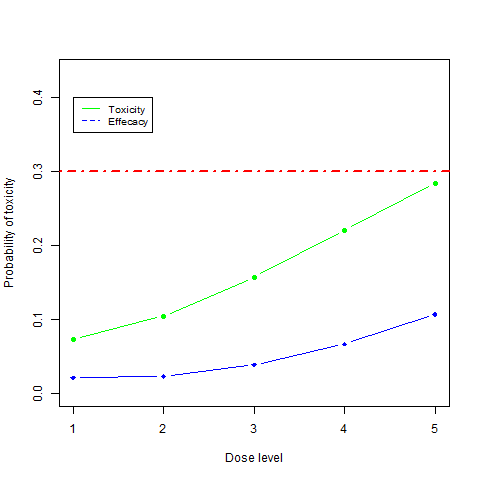
plot(1:ndoses, fit$prob_tox, type="b", pch=19, xlab="Dose level", ylab="Probability of toxicity", ylim=c(0,max(fit$prob_tox) + 0.15), col="green")
points(1:ndoses,fit$prob_eff, type="b", pch=18, col="blue")
abline(h=0.3, lwd=2, lty=4, col = "red")
legend(1, 0.4, legend=c("Toxicity", "Effecacy"),
col=c("green", "blue"), lty=1:2, cex=0.8)
this produce this figure
Thanks in advance