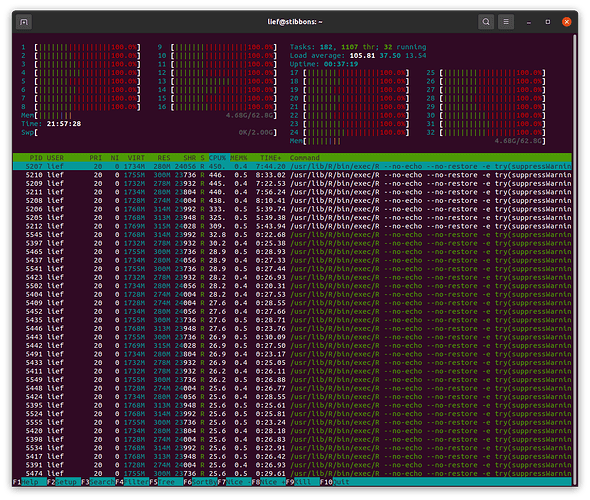
I am trying to use furrr::pmap to run a set of 216 models. The code runs fine on my 2015 macbook pro laptop (macos 10.15.7, 8gb of ram, 4 cores), and it works on my colleagues windows 10 pc. However, when I try to run it on my ubuntu machine (v 20.04, 64 gb ram, 32 cores) it gets progressively slower at running the models. Each model gets saved to disk, and I can see dramatic increases in the amount of time between file writes. See the attached screenshot - I killed the session around 9:30 pm when the last model output had been saved at 7:30
plan multicore vs multisession shows same behavior
I've observed the same behavior using both plan(multisession, workers = availableCores()) called from RStudio and using plan(multicore) and calling from an R session running in the terminal. I've also tried reducing the number of workers from 32 to 8 with no luck.
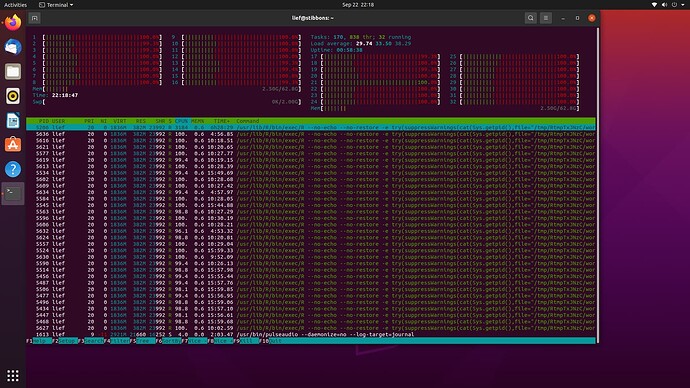
htop processor analysis
Examining htop it looks like something isn't working correctly with the cores. This screenshot was taken when I set plan (multisession, workers = 8), but you can see activity on all 32 cores. You can also see that in addition to the 8 main sessions of R there many other R sessions that are also occurring. I'm wondering if something is causing extra sessions to spin up and not close down that is bogging down the whole system over time.
memory
The data set that is being analyzed in the model is pretty small at just 2.9Mb. I don't see any indications that I'm running out of memory.
I'm a bit of a novice with linux computing and parallelization and would appreciate any suggestions of things to investigate.
Code
This is my invocation of future_pmap where multiverse_run is a custom function that runs the statistical model, saves the full output to disk and returns some summary statistics. multiverse_spec is a tibble that lists model parameters, and analysis_dat is the above mentioned 2.9Mb dataset. It takes ~1 minute to run per model.
multiverse_output <- multiverse_spec %>%
future_pmap(., possibly(multiverse_run, otherwise = "error"),
data = analysisdat,
.options = furrr_options(seed = TRUE),
.progress = T)
The full code is available here, I haven't had luck creating a minimal reproducible example of the code. But, given that it works fine on mac and windows I don't think the modelling code is the problem.