Hello from a new R user. I want to perform the statistical analysis of my data through R, but been facing a lot of problems. I want to perform a one stage dose-response with dosresmeta package. My data set include only 6 studies and each one only have two doses(low and high). For the studies I have the cases, control, n, logrr and se. However, when performing the codes I face problems like "must have positive length" or"Error in dosresmeta::dosresmeta(logRR ~ rcs(dose, 3), id = id, type = "cc", : The dimension of Slist does not match the data". Please I ask help on how should my dataset be organized and what are the proper codes for it. Thanks!
Hi @Linda1
Welcome to the Community Forum.
To get help you will need to follow the posting guide. That means you'll need to show us the data you are trying to analyse (anonymised if required) plus the code that you have tried already plus any output.
Thanks for the reply, here is the code I want to use and my data.
Slist <- split(data, data$id) %>% lapply(function(x) {
if (any(is.na(x$cases) | is.na(x$n))) {diag(x$se_logRR[x$se_logRR != 0 & !is.na(x$se_logRR)]^2,
nrow = sum(x$se_logRR != 0 & !is.na(x$se_logRR)))} else { matrix(x$se_logRR^2, nrow = length(x$se_logRR), ncol = length(x$se_logRR)) } })
print(Slist)
Up to this code, everything seems fine, however when I want to apply the formula:
spl_noexcl <- dosresmeta::dosresmeta(
logRR ~ rcs(dose, 3), # Restricted cubic splines with 3 knots
id = id,
type = "cc",
cases = cases,
n = n,
se = se_logRR,
data = data,
Slist = Slist,
covariance = "user",
proc = "1stage" )
Appears : Error in dosresmeta::dosresmeta(logRR ~ rcs(dose, 3), id = id, type = "cc", :
The dimension of Slist does not match the data
Hi @Linda1
We can't run and test your code without the data in text format.
Please post the output from dput(data).
Understand, sorry for the troublesome. Still in babysteps with R.
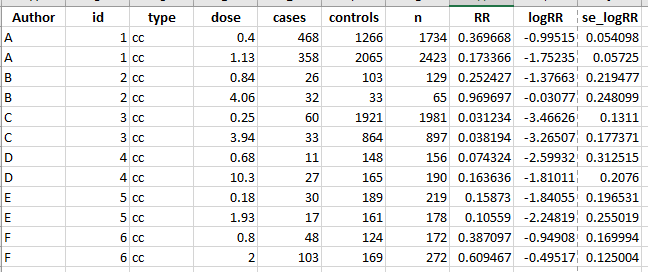
structure(list(Author = c("A", "A", "B", "B", "C", "C", "D",
"D", "E", "E", "F", "F"), id = c(1, 1, 2, 2, 3, 3, 4, 4, 5, 5,
6, 6), type = c("cc", "cc", "cc", "cc", "cc", "cc", "cc", "cc",
"cc", "cc", "cc", "cc"), dose = c(0.4, 1.13, 0.84, 4.06, 0.25,
3.94, 0.68, 10.3, 0.18, 1.93, 0.8, 2), cases = c(468, 358, 26,
32, 60, 33, 11, 27, 30, 17, 48, 103), controls = c(1266, 2065,
103, 33, 1921, 864, 148, 165, 189, 161, 124, 169), n = c(1734,
2423, 129, 65, 1981, 897, 156, 190, 219, 178, 172, 272), RR = c(0.369668246445498,
0.173365617433414, 0.252427184466019, 0.96969696969697, 0.0312337324310255,
0.0381944444444444, 0.0743243243243243, 0.163636363636364, 0.158730158730159,
0.105590062111801, 0.387096774193548, 0.609467455621302), logRR = c(-0.995149306786475,
-1.75235251899443, -1.37663245020815, -0.0307716586667537, -3.46625660054646,
-3.26506520733758, -2.59931700096574, -1.81010860789625, -1.84054963339749,
-2.24819102092825, -0.949080554697146, -0.495169726693438), se_logRR = c(0.0540984431590593,
0.0572499571230942, 0.219477279748078, 0.248098831724437, 0.131100071982629,
0.177370904351412, 0.312515355888071, 0.207599718443073, 0.19653075745119,
0.255019037595212, 0.169993674771638, 0.125003590458)), row.names = c(NA,
-12L), class = c("tbl_df", "tbl", "data.frame"))
Hi @Linda1
Sorry, I cannot get your code to work as I am not familiar with dosresmeta. Looks like the covariance matrices are not being constructed correctly. Maybe you can contact a colleague who has experience with this type of analysis?
Also worth noting that your script needs to load three packages:
library(dosresmeta)
library(rms)
library(tidyverse)
Thanks for the reply. I will see how else I can find the solution. ![]()
This topic was automatically closed 90 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.