Hi all,
Yet again, stumped by something that seemed simple.
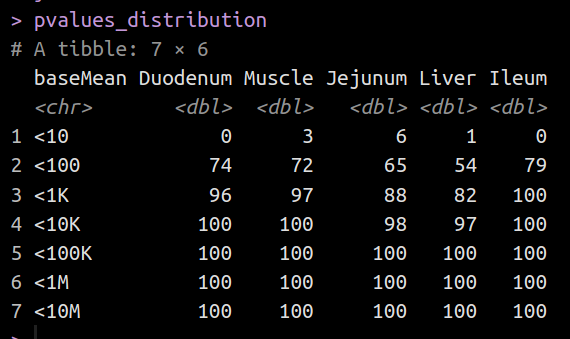
I have a list of deseqdataset objects (1 from each of 5 tissues). I am extracting some data from each tissue, and storing the data in 1 column (for each tissue) in a tibble. So the final tibble has 6 columns. 1 for each tissue, and the join_by key (baseMean). The problem is I can't figure out how to assign the tissue name to each column without hard coding it.
The column names I want:
# Initialize the tibble
> pvalues_distribution <- tibble(baseMean = c("<10", "<100", "<1K", "<10K", "<100K", "<1M", "<10M")) %>%
mutate(baseMean = factor(baseMean, baseMean)) # maintains row order
# Populate the tibble
> for (name in names(deseq2_results$T1)) {
# extract required columns + filter rows
res <- as_tibble(data.frame(deseq2_results$T1[[name]])) %>%
filter(pvalue < 0.1) %>%
filter(!between(log2FoldChange, -1.5, 1.5)) %>%
select(baseMean, pvalue, log2FoldChange)
# calculate % frequency distribution of pvalues
dist <- tibble(
x = c(10, 100, 1000, 10000, 100000, 1000000, 10000000),
baseMean = c("<10", "<100", "<1K", "<10K", "<100K", "<1M", "<10M")) %>%
rowwise() %>%
mutate(name = round(100 * (nrow(filter(res, baseMean < x)) / nrow(res)))) %>%
select(-1)
# join each tissue
pvalues_distribution <- left_join(pvalues_distribution, dist, join_by(baseMean))
}
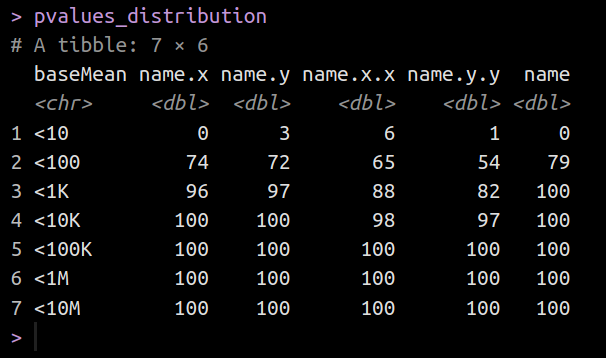
As you can see, the setting of the column name for each tissue when I call mutate() is interpreted as 'name' instead of the tissue it came from.
Thanks for the help
Kenneth