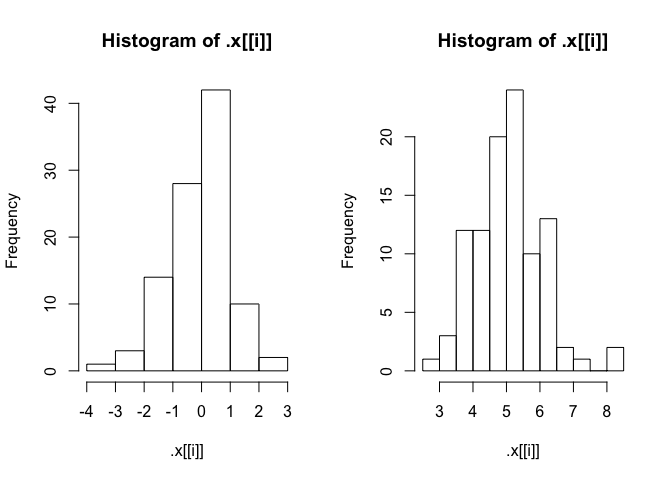
I have a problem when mapping the plot function over a list.
When iterating over the list for plotting I also get output in the console.
I do not want the output. I just want the plots. Just as it works when I use the function without
mapping: hist(rnorm(100))
I use purrr::map in the example but the same problem occurs when I use lapply()
Any suggestions?
library(dplyr)
# Setup plotting
par(mfrow =c(1,2))
# Create a list of data
list(a = rnorm(100), b = rnorm(100, 5)) %>%
# Map to histogram
purrr::map(hist)
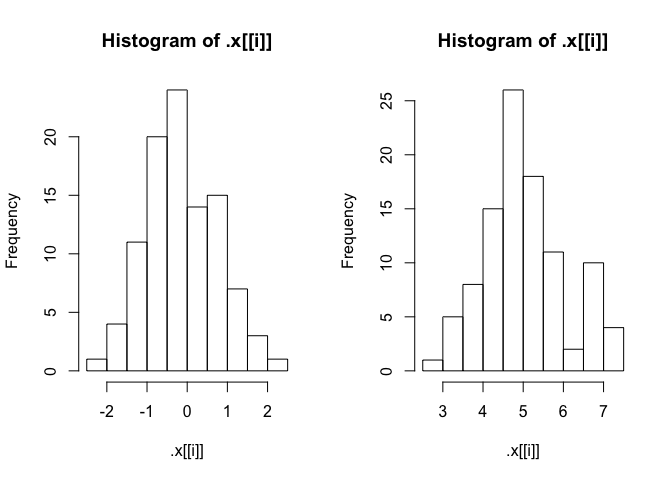
But then I get this "extra" output.
#> $a
#> $breaks
#> [1] -2.5 -2.0 -1.5 -1.0 -0.5 0.0 0.5 1.0 1.5 2.0 2.5
#>
#> $counts
#> [1] 1 4 11 20 24 14 15 7 3 1
#>
#> $density
#> [1] 0.02 0.08 0.22 0.40 0.48 0.28 0.30 0.14 0.06 0.02
#>
#> $mids
#> [1] -2.25 -1.75 -1.25 -0.75 -0.25 0.25 0.75 1.25 1.75 2.25
#>
#> $xname
#> [1] ".x[[i]]"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> $b
#> $breaks
#> [1] 2.5 3.0 3.5 4.0 4.5 5.0 5.5 6.0 6.5 7.0 7.5
#>
#> $counts
#> [1] 1 5 8 15 26 18 11 2 10 4
#>
#> $density
#> [1] 0.02 0.10 0.16 0.30 0.52 0.36 0.22 0.04 0.20 0.08
#>
#> $mids
#> [1] 2.75 3.25 3.75 4.25 4.75 5.25 5.75 6.25 6.75 7.25
#>
#> $xname
#> [1] ".x[[i]]"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
# Alternative for including title
# imap(~hist(.x, main = .y))
Created on 2019-05-31 by the reprex package (v0.3.0)