I'm working within hosted RStudio on a Linux EC2 server with 4 processors. In the past when I've used packages such as XGBoost or foreach, which use parallel processing, I have been able to watch the terminal and see all 4 processors light up within linux with top and pressing 1.
When I do that now here's what I see:
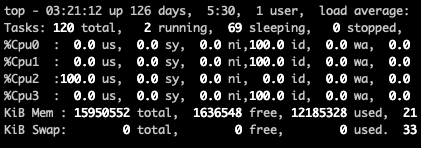
There's only one processor working. I expected to see multiple processors at work like I have in the past.
Here's my code block. Hoping someone can recognize what I'm not doing right here? This is my first time using furrr.
library(rsample)
library(pscl) # hurdle and zero inflated models
library(furrr) # parallel processing
plan(multicore) # also tried multiprocess but no change, only one processor seems to run
# otherwise error:
# Error in getGlobalsAndPackages(expr, envir = envir, tweak = tweakExpression, :
# The total size of the 9 globals that need to be exported for the future expression (‘{; ...future.f.env <- environment(...future.f); if (!is.null(...future.f.env$`~`)) {; if (is_bad_rlang_tilde(...future.f.env$`~`)) {; ...future.f.env$`~` <- base::`~`; }; ...; .out; }); }’) is 1.76 GiB. This exceeds the maximum allowed size of 1.46 GiB (option 'future.globals.maxSize'). The three largest globals are ‘...future.x_ii’ (1.76 GiB of class ‘list’), ‘is_bad_rlang_tilde’ (15.05 KiB of class ‘function’) and ‘...future.map’ (6.61 KiB of class ‘function’).
options(future.globals.maxSize = 2000 * 1024^2)
# create train test split
set.seed(42)
pdata_split <- initial_split(pdata, 0.9)
training_data <- training(pdata_split)
testing_data <- testing(pdata_split)
# cross validation folds
pdata_cv <- vfold_cv(training_data, 5, strata = spend_30d) %>%
# create training and validation sets within each fold
mutate(train = map(splits, ~training(.x)),
test = map(splits, ~testing(.x))) %>%
# hurdle model for each fold
mutate(hurdle_model = furrr::future_map(train,
~hurdle(formula = spend_30d ~.,
data = .x,
dist = "negbin")))