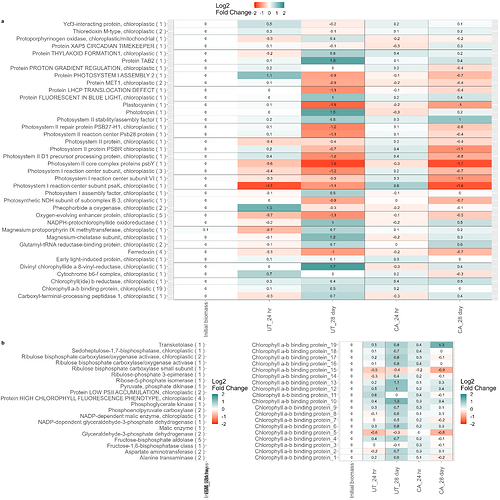
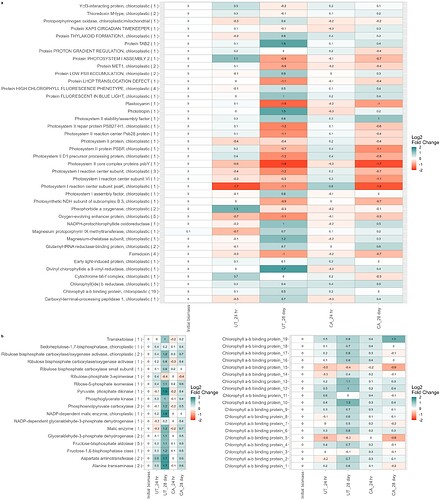
So I have three plots (information on these below) and I want to arrange them so that the first plot is the only one in the first row, and the other two are in the bottom row, and then try to scale them to fit everything in a readable format.
data:
> head(dat.light)
# A tibble: 6 × 4
title Role sample value
<chr> <chr> <chr> <dbl>
1 Carboxyl-terminal-processing peptidase 1, chloroplastic ( 1 ) PSII processing Initial biomass -0.00215
2 Carboxyl-terminal-processing peptidase 1, chloroplastic ( 1 ) PSII processing UT_24 hr -0.282
3 Carboxyl-terminal-processing peptidase 1, chloroplastic ( 1 ) PSII processing UT_28 day 0.676
4 Carboxyl-terminal-processing peptidase 1, chloroplastic ( 1 ) PSII processing CA_24 hr -0.256
5 Carboxyl-terminal-processing peptidase 1, chloroplastic ( 1 ) PSII processing CA_28 day 0.358
6 Chlorophyll a-b binding protein, chloroplastic ( 19 ) Light-Harvesting Complex Initial biomass -0.00148
> head(dat.dark)
# A tibble: 6 × 4
title Role sample value
<chr> <chr> <chr> <dbl>
1 Protein HIGH CHLOROPHYLL FLUORESCENCE PHENOTYPE, chloroplastic ( 4 ) PSII Assembly Initial biomass 0.0085
2 Protein HIGH CHLOROPHYLL FLUORESCENCE PHENOTYPE, chloroplastic ( 4 ) PSII Assembly UT_24 hr -0.0875
3 Protein HIGH CHLOROPHYLL FLUORESCENCE PHENOTYPE, chloroplastic ( 4 ) PSII Assembly UT_28 day 0.484
4 Protein HIGH CHLOROPHYLL FLUORESCENCE PHENOTYPE, chloroplastic ( 4 ) PSII Assembly CA_24 hr 0.0676
5 Protein HIGH CHLOROPHYLL FLUORESCENCE PHENOTYPE, chloroplastic ( 4 ) PSII Assembly CA_28 day 0.441
6 Protein LOW PSII ACCUMULATION, chloroplastic ( 2 ) PSII Assembly Initial biomass -0.0006
> head(cbp_longer)
# A tibble: 6 × 3
Protein sample value
<chr> <chr> <dbl>
1 Chlorophyll a-b binding protein_1 Initial biomass -0.00125
2 Chlorophyll a-b binding protein_1 UT_24 hr 0.198
3 Chlorophyll a-b binding protein_1 UT_28 day 0.599
4 Chlorophyll a-b binding protein_1 CA_24 hr 0.148
5 Chlorophyll a-b binding protein_1 CA_28 day -0.193
6 Chlorophyll a-b binding protein_2 Initial biomass 0.016
Code for plots;
plot_light<-ggplot(dat.light, aes(x = factor(sample,level=level_order), y=title,fill=value)) +
geom_tile(color = "black")+
scale_fill_gradient2(limits=c(-2,2),low = "red",
mid = "white",
high = "cyan4")+
guides(fill=guide_colourbar(title='Log2 \nFold Change'))+
theme(text=element_text(size=18),axis.text=element_text(size=16),axis.text.x=element_text(angle=90,vjust=0.5),axis.title=element_blank(),plot.title=element_text(size=18,hjust=0.5))+
geom_text(aes(label=round(value,1)))
photo_dark<-ggplot(dat.dark, aes(x = factor(sample,level=level_order), y=title,fill=value)) +
geom_tile(color = "black")+
scale_fill_gradient2(limits=c(-2,2),low = "red",
mid = "white",
high = "cyan4")+
# coord_fixed(ratio=0.2)+
guides(fill=guide_colourbar(title='Log2 \nFold Change'))+
theme(text=element_text(size=18),axis.text=element_text(size=16),axis.text.x=element_text(angle=90,vjust=0.5),axis.title=element_blank(),plot.title=element_text(size=18,hjust=0.5))+
geom_text(aes(label=round(value,1)))
cbp_plot<-ggplot(cbp_longer, aes(x = factor(sample,level=level_order), y=factor(Protein,level=prot_level),fill=value)) +
geom_tile(color = "black")+
scale_fill_gradient2(limits=c(-2,2),low = "red",
mid = "white",
high = "cyan4")+
guides(fill=guide_colourbar(title='Log2 \nFold Change'))+
theme(text=element_text(size=18),axis.text=element_text(size=16),axis.text.x=element_text(angle=90,vjust=0.5),axis.title=element_blank(),plot.title=element_text(size=18,hjust=0.5))+
geom_text(aes(label=round(value,1)))
I'm using ggarrange() to try to format them, but I can't figure out how to get the photo_light centered and alone in the first row, and put the other two on the bottom row. Also, any tips on formatting for these is greatly appreciated as this is new to me.
ggarrange(plot_light,photo_dark,cbp_plot,ncol=2,nrow=2,heights=c(2,0.7),labels="auto",common.legend=TRUE)