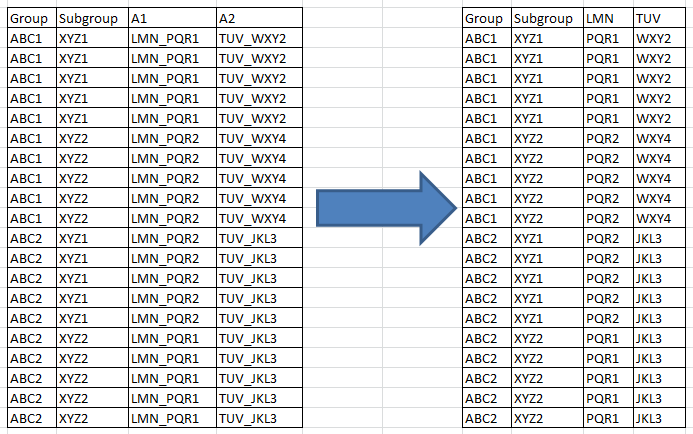
I have tidyr code to get me a table as shown below.on the left. I am trying to get to output on the right which can split/separate columns and use first column from the pair as column name for the 2nd column.
Hi!
To help us help you, could you please prepare a reproducible example (reprex) illustrating your issue? Please have a look at this guide, to see how to create one:
I dont really have any code for it yet.
But this is what I would think of starting with...
library(tidyr)
dt <- data.frame(Group = c(rep("ABC1", 10),rep("ABC2", 10)),
SubGroup = c(rep("XYZ1", 5), rep("XYZ2", 5), rep("XYZ1", 5), rep("XYZ2", 5)),
A1 = c(rep("LMN_PQR1", 5), rep("LMN_PQR2", 10), rep("LMN_PQR1", 5)),
A2 = c(rep("TUV_WXY2", 5), rep("TUV_WXY4", 5), rep("TUV_JKL3", 10)))
dt %>%
separate(A1, c("Col1", "Col2"), sep = "_", extra = "merge") %>%
separate(A2, c("Col3", "Col4"), sep = "_", extra = "merge")
The columns to be separated are variable and can range from several 10s to 100s. The first two are fixed.
So I am looking for a generalized loop that can take dt[,-1:-2] as input, separate them and for each separation make unique value from first column as header for 2nd col.
I'm sure there is a more elegant way but in the mean while this works
library(tidyverse)
dt <- data.frame(Group = c(rep("ABC1", 10),rep("ABC2", 10)),
SubGroup = c(rep("XYZ1", 5), rep("XYZ2", 5), rep("XYZ1", 5), rep("XYZ2", 5)),
A1 = c(rep("LMN_PQR1", 5), rep("LMN_PQR2", 10), rep("LMN_PQR1", 5)),
A2 = c(rep("TUV_WXY2", 5), rep("TUV_WXY4", 5), rep("TUV_JKL3", 10)))
names <- dt %>%
select(-Group, -SubGroup) %>%
head(1) %>%
unlist() %>%
str_extract(".+(?=_)")
dt %>%
transmute_at(vars(-Group, -SubGroup), ~str_extract(., "(?<=_).+"), ) %>%
rename_all(~names) %>%
bind_cols(dt[,1:2]) %>%
select(Group, SubGroup, everything())
#> Group SubGroup LMN TUV
#> 1 ABC1 XYZ1 PQR1 WXY2
#> 2 ABC1 XYZ1 PQR1 WXY2
#> 3 ABC1 XYZ1 PQR1 WXY2
#> 4 ABC1 XYZ1 PQR1 WXY2
#> 5 ABC1 XYZ1 PQR1 WXY2
#> 6 ABC1 XYZ2 PQR2 WXY4
#> 7 ABC1 XYZ2 PQR2 WXY4
#> 8 ABC1 XYZ2 PQR2 WXY4
#> 9 ABC1 XYZ2 PQR2 WXY4
#> 10 ABC1 XYZ2 PQR2 WXY4
#> 11 ABC2 XYZ1 PQR2 JKL3
#> 12 ABC2 XYZ1 PQR2 JKL3
#> 13 ABC2 XYZ1 PQR2 JKL3
#> 14 ABC2 XYZ1 PQR2 JKL3
#> 15 ABC2 XYZ1 PQR2 JKL3
#> 16 ABC2 XYZ2 PQR1 JKL3
#> 17 ABC2 XYZ2 PQR1 JKL3
#> 18 ABC2 XYZ2 PQR1 JKL3
#> 19 ABC2 XYZ2 PQR1 JKL3
#> 20 ABC2 XYZ2 PQR1 JKL3
Created on 2019-08-30 by the reprex package (v0.3.0.9000)
Here's a way you can do that:
The script is more wordy than it needs to be to make it easier to follow.
Since I qualified the tidyverse functions you may not need to load them, or you can replace the 4 library calls with a single library(tidyverse) to load all of them at once.
library(tidyr)
library(dplyr)
library(stringr)
library(purr)
dt <- data.frame(
Group = c(rep("ABC1", 10), rep("ABC2", 10)),
SubGroup = c(rep("XYZ1", 5), rep("XYZ2", 5), rep("XYZ1", 5), rep("XYZ2", 5)),
A1 = c(rep("LMN_PQR1", 5), rep("LMN_PQR2", 10), rep("LMN_PQR1", 5)),
A2 = c(rep("TUV_WXY2", 5), rep("TUV_WXY4", 5), rep("TUV_JKL3", 10)),
stringsAsFactors = FALSE
)
- Define a function that will take a column name from a data.frame, get the content of the first row, split by "_", take the first character vector result as new column name, and then proceed to separate the column (default is splitting by non_alpha character, as such it will split at the "_"). NA for the first column name will skip creating the column. [If you're in the same environment, you may be able to remove the second parameter, dframe and use dt directly. In that case also lose the corresponding argument to map_dfc.]
split_column <- function(col, dframe) {
row1 <- dframe[1, col]
new_col_name <- stringr::str_split_fixed(row1, "_", n = 2)[, 1]
new_col <- dframe %>%
dplyr::select(col) %>%
tidyr::separate(col, c(NA, new_col_name))
}
- Build a list of column names without the first 2:
cols <- colnames(dt)[-1:-2]
- Then run the split_column function over all remaining columns with map_dfc, which returns a data frame with the combined columns:
new_cols <- purrr::map_dfc(cols, split_column, dt)
- Add in the original first 2 columns to create the modified dt:
new_dt <- dplyr::bind_cols(dt[1:2], new_cols)
And here's the result:
Group SubGroup LMN TUV
1 ABC1 XYZ1 PQR1 WXY2
2 ABC1 XYZ1 PQR1 WXY2
3 ABC1 XYZ1 PQR1 WXY2
4 ABC1 XYZ1 PQR1 WXY2
5 ABC1 XYZ1 PQR1 WXY2
6 ABC1 XYZ2 PQR2 WXY4
7 ABC1 XYZ2 PQR2 WXY4
8 ABC1 XYZ2 PQR2 WXY4
9 ABC1 XYZ2 PQR2 WXY4
10 ABC1 XYZ2 PQR2 WXY4
11 ABC2 XYZ1 PQR2 JKL3
12 ABC2 XYZ1 PQR2 JKL3
13 ABC2 XYZ1 PQR2 JKL3
14 ABC2 XYZ1 PQR2 JKL3
15 ABC2 XYZ1 PQR2 JKL3
16 ABC2 XYZ2 PQR1 JKL3
17 ABC2 XYZ2 PQR1 JKL3
18 ABC2 XYZ2 PQR1 JKL3
19 ABC2 XYZ2 PQR1 JKL3
20 ABC2 XYZ2 PQR1 JKL3
Voilà
Here's the script that I suggest that use rename_at function:
library(tidyverse)
dt <- data.frame(Group = c(rep("ABC1", 10),rep("ABC2", 10)),
SubGroup = c(rep("XYZ1", 5), rep("XYZ2", 5), rep("XYZ1", 5), rep("XYZ2", 5)),
A1 = c(rep("LMN_PQR1", 5), rep("LMN_PQR2", 10), rep("LMN_PQR1", 5)),
A2 = c(rep("TUV_WXY2", 5), rep("TUV_WXY4", 5), rep("TUV_JKL3", 10)))
dt %>%
rename_at(vars(-Group, -SubGroup), funs(map_chr(~str_split(.x[1], "_")[[1]][1]), .args = list(.x = select(., -Group, -SubGroup)))) %>%
mutate_at(vars(-Group, -SubGroup), ~str_split(., "_", simplify = TRUE)[, 2])
This topic was automatically closed 21 days after the last reply. New replies are no longer allowed.