Overview
I have produced four models using the tidymodels package with the data frame FID (see below):
- General Linear Model
- Bagged Tree
- Random Forest
- Boosted Trees
The data frame contains three predictors :
- Year (numeric)
- Month (Factor)
- Days (numeric)
The dependent variable is Frequency (numeric)
Aim
My aim is to undertake model predictions to extract the class and probability values for all fitted models , which have all undergone 10 fold cross-validation .
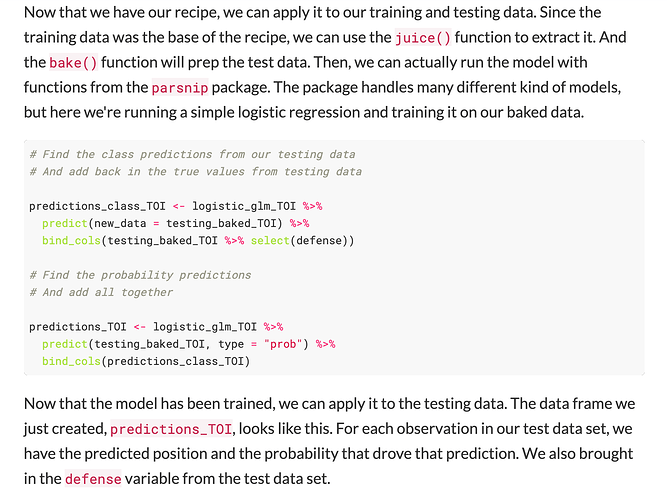
I am attempting to use the functions prep(), juice(), and bake() in order to generate the correct data objects for model predictions objects by following this tutorial below.
Tutorial (see screenshots below)
https://meghan.rbind.io/post/tidymodels-intro/
After producing the model prediction values (i.e class and probability) for all four models, the ultimate aim is to produce confusion matrices and receiver operating curves (ROC) to evaluate all models. Therefore, I need to bind the true values from the testing data, with the class and probability columns extracted from these model predictions.
Issue
I am trying to run the predict() function to produce the class and probability values from the tutorial (see screenshots below and the link above), but I am experiencing this error message below.
**Error Messages**
##Class prediction object
Error in UseMethod("predict") :
no applicable method for 'predict' applied to an object of class "c('tbl_df', 'tbl', 'data.frame')"
##Prob
Error in UseMethod("predict") :
no applicable method for 'predict' applied to an object of class "c('resample_results', 'tune_results', 'tbl_df', 'tbl', 'data.frame')"
If anyone is able to help, I would be deeply appreciative
Many thanks in advance.
Screen-shots from the tutorial
R-code
##################################################
##Model Prediction
###################################################
##Open the tidymodels package
library(tidymodels)
library(tidyverse)
library(glmnet)
library(parsnip)
library(rpart)
library(tidyverse) # manipulating data
library(skimr) # data visualization
library(baguette) # bagged trees
library(future) # parallel processing & decrease computation time
library(xgboost) # boosted trees
library(ranger)
library(yardstick)
library(purrr)
library(forcats)
###########################################################
#split this single dataset into two: a training set and a testing set
data_split <- initial_split(FID)
# Create data frames for the two sets:
train_data <- training(data_split)
test_data <- testing(data_split)
# resample the data with 10-fold cross-validation (10-fold by default)
cv <- vfold_cv(train_data, v=10)
###########################################################
##Produce the recipe
rec <- recipe(Frequency ~ ., data = FID) %>%
step_nzv(all_predictors(), freq_cut = 0, unique_cut = 0) %>% # remove variables with zero variances
step_novel(all_nominal()) %>% # prepares test data to handle previously unseen factor levels
step_medianimpute(all_numeric(), -all_outcomes(), -has_role("id vars")) %>% # replaces missing numeric observations with the median
step_dummy(all_nominal(), -has_role("id vars")) # dummy codes categorical variables
###########################################################
##Create Models
###########################################################
##########################################################
##General Linear Models
#########################################################
##glm
mod_glm<-linear_reg(mode="regression",
penalty = 0.1,
mixture = 1) %>%
set_engine("glmnet")
##Create workflow
wflow_glm <- workflow() %>%
add_recipe(rec) %>%
add_model(mod_glm)
##Fit the model
plan(multisession)
fit_glm <- fit_resamples(
wflow_glm,
cv,
metrics = metric_set(rmse, rsq),
control = control_resamples(save_pred = TRUE,
extract = function(x) extract_model(x)))
##########################################################
##Bagged Trees
##########################################################
#####Bagged Trees
mod_bag <- bag_tree() %>%
set_mode("regression") %>%
set_engine("rpart", times = 10) #10 bootstrap resamples
##Create workflow
wflow_bag <- workflow() %>%
add_recipe(rec) %>%
add_model(mod_bag)
##Fit the model
plan(multisession)
fit_bag <- fit_resamples(
wflow_bag,
cv,
metrics = metric_set(rmse, rsq),
control = control_resamples(save_pred = TRUE,
extract = function(x) extract_model(x)))
###################################################
##Random forests
###################################################
mod_rf <-rand_forest(trees = 1e3) %>%
set_engine("ranger",
num.threads = parallel::detectCores(),
importance = "permutation",
verbose = TRUE) %>%
set_mode("regression")
##Create Workflow
wflow_rf <- workflow() %>%
add_model(mod_rf) %>%
add_recipe(rec)
##Fit the model
plan(multisession)
fit_rf<-fit_resamples(
wflow_rf,
cv,
metrics = metric_set(rmse, rsq),
control = control_resamples(save_pred = TRUE,
extract = function(x) extract_model(x)))
############################################################
##Boosted Trees
############################################################
mod_boost <- boost_tree() %>%
set_engine("xgboost", nthreads = parallel::detectCores()) %>%
set_mode("regression")
##Create Workflow
wflow_boost <- workflow() %>%
add_recipe(rec) %>%
add_model(mod_boost)
##Fit model
plan(multisession)
fit_boost <-fit_resamples(
wflow_boost,
cv,
metrics = metric_set(rmse, rsq),
control = control_resamples(save_pred = TRUE,
extract = function(x) extract_model(x)))
##################################################
##Prep the models for model prediction
##################################################
# Extract our prepped training data
# and "bake" our testing data
prep<-prep(rec)
training_baked<-juice(prep)
testing_baked <- prep %>% bake(test_data)
# Run the model with our training data
# Find the class predictions from our testing data
# And add back in the true values from testing data
predictions_class <- %>% fit_glm %>%
predict(new_data = testing_baked) %>%
bind_cols(testing_baked %>% dplyr::select(Frequency))
##Error message
Error in UseMethod("predict") :
no applicable method for 'predict' applied to an object of class "c('tbl_df', 'tbl', 'data.frame')"
# Find the probability predictions
# And add all together
predictions_Prob <- fit_glm %>%
predict(testing_baked, type = "prob") %>%
bind_cols(predictions_class)
##Error message
Error in UseMethod("predict") :
no applicable method for 'predict' applied to an object of class
"c('resample_results', 'tune_results', 'tbl_df', 'tbl', 'data.frame')"
Data frame - FID
structure(list(Year = c(2015, 2015, 2015, 2015, 2015, 2015, 2015,
2015, 2015, 2015, 2015, 2015, 2016, 2016, 2016, 2016, 2016, 2016,
2016, 2016, 2016, 2016, 2016, 2016, 2017, 2017, 2017, 2017, 2017,
2017, 2017, 2017, 2017, 2017, 2017, 2017), Month = structure(c(1L,
2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L,
5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L,
8L, 9L, 10L, 11L, 12L), .Label = c("January", "February", "March",
"April", "May", "June", "July", "August", "September", "October",
"November", "December"), class = "factor"), Frequency = c(36,
28, 39, 46, 5, 0, 0, 22, 10, 15, 8, 33, 33, 29, 31, 23, 8, 9,
7, 40, 41, 41, 30, 30, 44, 37, 41, 42, 20, 0, 7, 27, 35, 27,
43, 38), Days = c(31, 28, 31, 30, 6, 0, 0, 29, 15,
29, 29, 31, 31, 29, 30, 30, 7, 0, 7, 30, 30, 31, 30, 27, 31,
28, 30, 30, 21, 0, 7, 26, 29, 27, 29, 29)), row.names = c(NA,
-36L), class = "data.frame")