I was following along with David Robinson's recent Sliced Competition Lap 2 on Youtube when I ran into an error using augment on the fitted workflow with the training or test data. Not sure what the problem is and haven't yet found a solution that I understand. I've created my first reprex below which yields a similar error.
It's a similar error message to the one posted here: tidymodels: error when predicting on new data with xgboost model - #2 by julia
Using Julia's reprex I get the error when piping the fitted workflow into augment() with training or test data.
library(tidymodels)
#> Registered S3 method overwritten by 'tune':
#> method from
#> required_pkgs.model_spec parsnip
library(reprex)
data("hpc_data")
xgb_spec <- boost_tree(
trees = 1000,
tree_depth = tune(),
min_n = tune()
) %>%
set_engine("xgboost") %>%
set_mode("classification")
spl <- initial_split(hpc_data)
training <- training(spl)
testing <- testing(spl)
hpc_folds <- vfold_cv(v = 5, training, strata = class)
xgb_grid <- grid_latin_hypercube(
tree_depth(),
min_n(),
size = 5
)
xgb_wf <- workflow() %>%
add_formula(class ~ .) %>%
add_model(xgb_spec)
xgb_wf
#> ══ Workflow ════════════════════════════════════════════════════════════════════
#> Preprocessor: Formula
#> Model: boost_tree()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> class ~ .
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> Boosted Tree Model Specification (classification)
#>
#> Main Arguments:
#> trees = 1000
#> min_n = tune()
#> tree_depth = tune()
#>
#> Computational engine: xgboost
# Tune workflow ----
doParallel::registerDoParallel()
set.seed(123)
xgb_res <- tune_grid(
xgb_wf,
resamples = hpc_folds,
grid = xgb_grid
)
xgb_res
#> # Tuning results
#> # 5-fold cross-validation using stratification
#> # A tibble: 5 x 4
#> splits id .metrics .notes
#> <list> <chr> <list> <list>
#> 1 <split [2597/651]> Fold1 <tibble [10 × 6]> <tibble [0 × 1]>
#> 2 <split [2598/650]> Fold2 <tibble [10 × 6]> <tibble [0 × 1]>
#> 3 <split [2598/650]> Fold3 <tibble [10 × 6]> <tibble [0 × 1]>
#> 4 <split [2599/649]> Fold4 <tibble [10 × 6]> <tibble [0 × 1]>
#> 5 <split [2600/648]> Fold5 <tibble [10 × 6]> <tibble [0 × 1]>
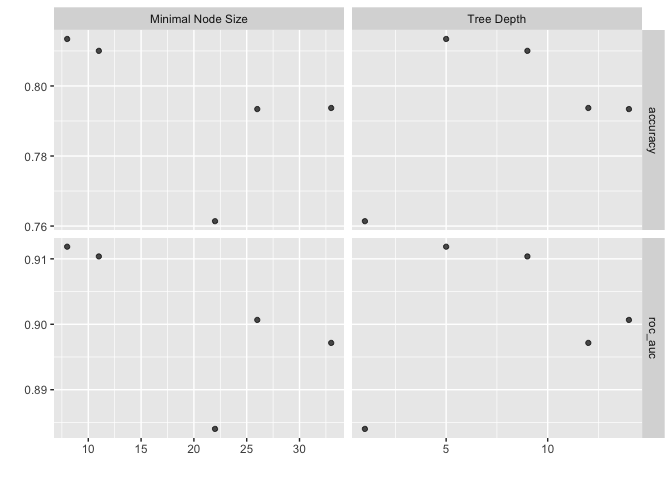
xgb_res %>% autoplot()
# Fit the best workflow to the training data ----
trained_wf <- xgb_wf %>%
finalize_workflow(
select_best(xgb_res, "roc_auc")
) %>%
fit(training)
#> [06:18:40] WARNING: amalgamation/../src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'multi:softprob' was changed from 'merror' to 'mlogloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
trained_wf
#> ══ Workflow [trained] ══════════════════════════════════════════════════════════
#> Preprocessor: Formula
#> Model: boost_tree()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> class ~ .
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> ##### xgb.Booster
#> raw: 4.9 Mb
#> call:
#> xgboost::xgb.train(params = list(eta = 0.3, max_depth = 5L, gamma = 0,
#> colsample_bytree = 1, colsample_bynode = 1, min_child_weight = 8L,
#> subsample = 1, objective = "multi:softprob"), data = x$data,
#> nrounds = 1000, watchlist = x$watchlist, verbose = 0, num_class = 4L,
#> nthread = 1)
#> params (as set within xgb.train):
#> eta = "0.3", max_depth = "5", gamma = "0", colsample_bytree = "1", colsample_bynode = "1", min_child_weight = "8", subsample = "1", objective = "multi:softprob", num_class = "4", nthread = "1", validate_parameters = "TRUE"
#> xgb.attributes:
#> niter
#> callbacks:
#> cb.evaluation.log()
#> # of features: 26
#> niter: 1000
#> nfeatures : 26
#> evaluation_log:
#> iter training_mlogloss
#> 1 1.128122
#> 2 0.977151
#> ---
#> 999 0.031266
#> 1000 0.031244
# Predict on brand new data ----
brand_new_data <- hpc_data[5, -8]
brand_new_data
#> # A tibble: 1 x 7
#> protocol compounds input_fields iterations num_pending hour day
#> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 E 100 82 20 0 10.4 Fri
predict(trained_wf, new_data = brand_new_data)
#> # A tibble: 1 x 1
#> .pred_class
#> <fct>
#> 1 VF
# Augment the testing data causes an error
trained_wf %>%
augment(testing)
#> Error in xgboost::xgb.DMatrix(data = newdata, missing = NA): 'data' has class 'character' and length 29241.
#> 'data' accepts either a numeric matrix or a single filename.
Created on 2021-07-26 by the [reprex package]