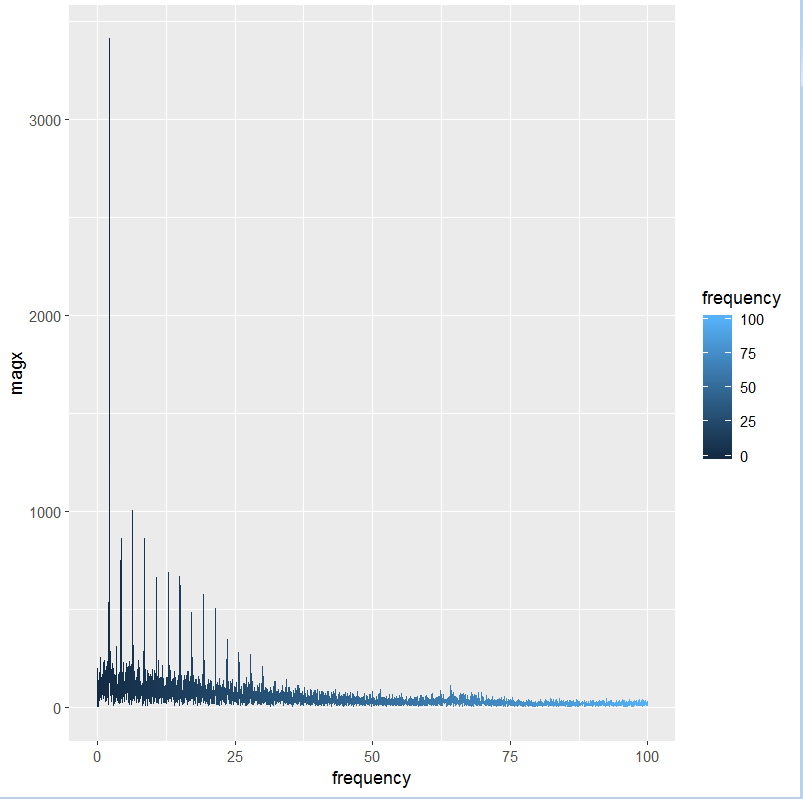
Below is a code in which the user uploads a csv file, the code then goes on to plot a fft plot, but as you will see when clicked on the fft button it does not plot the data points on the graph, if anyone could help me, i would be really grateful.
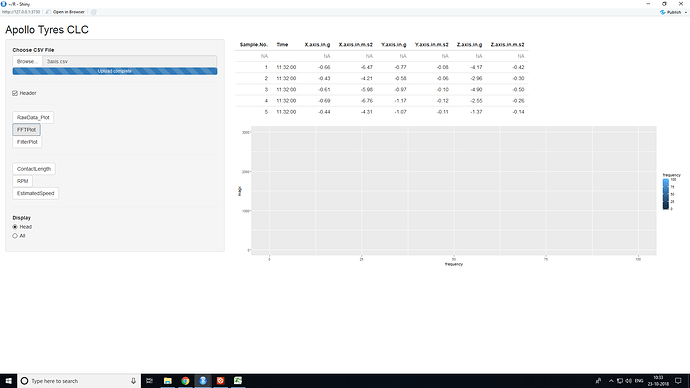
i will upload the image of the app,which after clicking on the FFTPLOT action button looks like this
find the screenshot PFA
Also the csv input file
apparently i cant upload it ![]()
library(ggplot2)
ui <- fluidPage(
titlePanel("Apollo Tyres CLC "),
sidebarLayout(
sidebarPanel(
fileInput("file1", "Choose CSV File",
multiple = FALSE,
accept = c("text/csv",
"text/comma-separated-values,text/plain",
".csv")),
tags$hr(),
checkboxInput("header", "Header", TRUE),
tags$hr(),
actionButton("rawdata","RawData_Plot"),
br(),
actionButton("fftdata","FFTPlot"),
br(),
actionButton("filter","FilterPlot"),
br(),
tags$hr(),
actionButton("clc","ContactLength"),
br(),
actionButton("rpm","RPM"),
br(),
actionButton("speed","EstimatedSpeed"),
br(),
tags$hr(),
# Input: Select number of rows to display ----
radioButtons("disp", "Display",
choices = c(Head = "head",
All = "all"),
selected = "head")
),
# Main panel for displaying outputs ----
mainPanel(
# Output: Data file ----
tableOutput("contents"),
plotOutput("fftplot")
)
)
)
# Define server logic to read selected file ----
server <- function(input, output) {
output$contents <- renderTable({
# input$file1 will be NULL initially. After the user selects
# and uploads a file, head of that data file by default,
# or all rows if selected, will be shown.
req(input$file1)
# when reading semicolon separated files,
# having a comma separator causes `read.csv` to error
tryCatch(
{
df <- read.csv(input$file1$datapath,
header = TRUE)
},
error = function(e) {
# return a safeError if a parsing error occurs
stop(safeError(e))
}
)
if(input$disp == "head") {
return(head(df))
}
else {
return(df)
}
})
@@@@@@@@@@#######So the problem is in this code@@@@@2
fftplot<- eventReactive(input$fftdata, {
infile <- input$file1
data <- read.csv(infile$datapath)
data$X.axis.in.g = data$X.axis.in.g - mean(data$X.axis.in.g) ##DC offset removal##
fftx = fft(data$X.axis.in.g) ##takes FFT##
magx = Mod(fftx[1:(length(fftx)/2)]) ##magnitude of FFT##
frequency = seq(0,100,length.out=length(magx)) ##frequency Range##
fftdf = data.frame(magx,frequency) ##making a data frame of freq and mag##
aabc = ggplot(fftdf,aes(x=frequency,y=magx,color=frequency))
aabc+ geom_line()+coord_cartesian(ylim=c(0,3000)) +
geom_smooth(method="lm")
})
output$fftplot <- renderPlot({fftplot()})
@@@@@@@@@@##########@@@@@@@@@
}
# Create Shiny app ----
shinyApp(ui, server)``````
the warning message i am getting is
Warning: Removed 5941 rows containing non-finite values (stat_smooth).
Warning: Removed 5941 rows containing missing values (geom_path).