Dear Ms./Mr.,
If the text and codes below are not properly displayed, you can download the Word file, which has exactly the same content in the link that is at the end of this message.
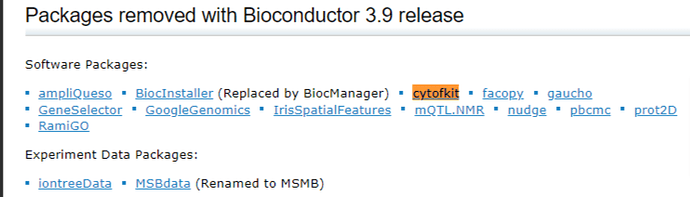
I am a cytometrist and microscopist at Izmir Institute of Technology, Integrated Research Center. I operate a cytometer and fluorescence microscope to acquire data from our researchers’ samples and then give them analyzed data. I need to dive into bulk data to present all of the information in the sample. To do this, I have recently been trying to develop my skills in R, RStudio, and Cytofkit package for cytometry (flow, spectral, mass, imaging mass cytometry) data analysis. I am new in this area and must learn R for data analysis.
May I request help from you or one of your assistants who is competent in R, RStudio, and R packages with the installation issues caused by “version differences” between R itself, R packages, and their dependencies? I really had to send this email to you anymore, because I could not run the package “Cytofkit” and find a solution on the internet.
It has been nearly 2 weeks since I have been trying to solve the errors and warnings. I am in a vicious circle and cannot move forward anymore. This is why I needed help from an expert competent in R programming.
I tried to use the renv package, but I could not move forward because I am not competent in R.
I explained the errors that I faced during the installation in the link below.
I have two computers, both operating Windows 10, 64-bit.
I installed R 3.5.2 on the desktop (Windows in English) and R 3.5.0 on the laptop (Windows in Turkish).
I know that you are very busy with your work, but may you (or your assistant) help me by informing me how to install consistent versions of R, RStudio, and packages/dependencies (for example, Cytofkit, ggplot2, Rtools, devtools, plyr, shiny, GUI)?
I guess I will need exact web page links "in an orderly manner" to download the correct versions of everything.
I would appreciate it if you could advise me or forward this email to one of your assistants or experts.
I really appreciate any help you can provide.
Kind Regards
Please find the technical details in the link below;
OR
Download the image and zoom it by right-clicking and "save image as"
Kind Regards
System Information:
- RStudio Edition: Desktop Latest
- RStudio Version: Desktop Latest
- OS Version: Windows 10
- R Version: 5.3.0 and 5.3.2
sessionInfo():
Referred here from support.rstudio.com