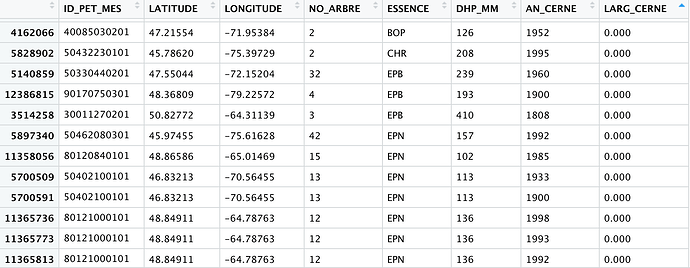
Hi there ! I've got a very very large data set (over 13 million entries). This data set represents tree ring widths (LARG_CERNE) collected for different trees (NO_ARBRE) at each site (ID_PET_MES). ESSENCE refers to the species the tree is. AN_CERNE is the year the tree ring width corresponds to.
Here is my issue: I want to organize or interrogate my data to be able to see at each site (ID_PET_MES) how many numbers of each species I have, so I can compare and select sites with the most trees of certain species. The only way I have managed this so far is to use the filter function to filter specific sites or specific species to create smaller data sets and then analyze them but I don't want to have to do this site by site as I have over 100 000 sites and it will take me too long.
For example in the image below. I want R to be able to tell me in the site with ID "40085030201" (first row), how many BOP trees do I have? How many EPN trees? How many CHR etc... And I want to do this for every site to be able to compare them.
Does anyone know how to do this?? Thank you