I have the following code:
model <- as.formula("y~z+x2+x4+x5+x6+x10+x11")
set.seed(12345)
suppressMessages(library(caret))
train.control <- trainControl(method="repeatedcv", number=10, repeats=3)
cv <- train(model,data=df2, method="glm", trControl=train.control)
cat("RMSE =",cv$results$RMSE,"\n")
cat("Rsquared =",cv$results$Rsquared,"\n")
cat("MAE =",cv$results$MAE,"\n")
The following statements result in NULL (each one)
cv$varImp
cv$aic
cv$deviance
When I see something listed in str(cv) am I to assume it can be available?
varImp, aic and deviance are included in the list.
The package documentation was sparse on this topic.
Hi, you can extract the AIC and deviance like this. I have used the iris dataset to make it reproducible.
model <- as.formula("Sepal.Length ~ Sepal.Width + Petal.Length")
set.seed(12345)
suppressMessages(library(caret))
train.control <- trainControl(method="repeatedcv", number=10, repeats=3)
cv <- train(model, data=iris, method="glm", trControl=train.control)
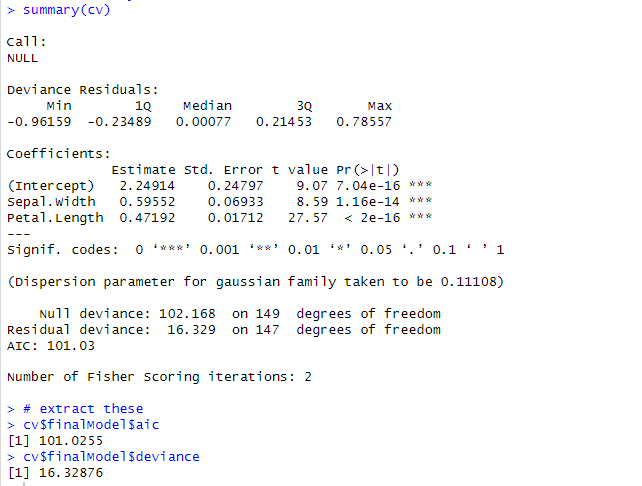
# view summary
summary(cv)
# extract these
cv$finalModel$aic
cv$finalModel$deviance
William,
Do you know why cv$ModelInfo$varImp does not work? Thanks for aic and deviance example. I tried to expand on that.
Mary
str(cv)
List of 24
$ method : chr "glm"
$ modelInfo :List of 15
..$ label : chr "Generalized Linear Model"
..$ library : NULL
..$ loop : NULL
..$ type : chr [1:2] "Regression" "Classification"
..$ parameters:'data.frame': 1 obs. of 3 variables:
.. ..$ parameter: chr "parameter"
.. ..$ class : chr "character"
.. ..$ label : chr "parameter"
..$ grid :function (x, y, len = NULL, search = "grid")
..$ fit :function (x, y, wts, param, lev, last, classProbs, ...)
..$ predict :function (modelFit, newdata, submodels = NULL)
..$ prob :function (modelFit, newdata, submodels = NULL)
..$ varImp :function (object, ...)
cv$ModelInfo$varImp
Output
NULL
I think it is because varImp()is a function.
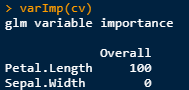
You can run varImp(cv) though.
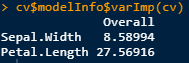
Though maybe it should be this: cv$modelInfo$varImp(cv)
William,
Note the definition of variable importance.
variable importance - mean decrease in node impurity (and not the mean decrease in accuracy)
To me this would imply that cv$modelInfo$varImp(cv) is the correct syntax. Getting the right stats out of a str function is tricky.
This is what I tried. Note the varImp values differ.
cv$modelInfo$varImp(cv) # 8-> 39.7
varImp(cv) # 100 -> 0
I also looked at:
library(caret)
library(randomForest)
varImpPlot(cv,type=2) # Note: This function only works for objects of class `randomForest'
From this I would conclude that cv$modelInfo$varImp(cv) is correct. Thank you replying.
1 Like


