Thank you for your patience.
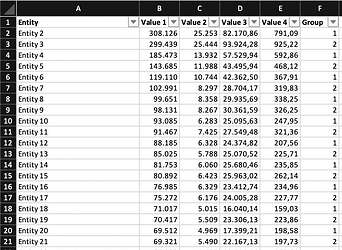
I tried renaming my data set for clarity. Now it is 'mdat'. I also added it to the first line (however I dont think it is necessary?)
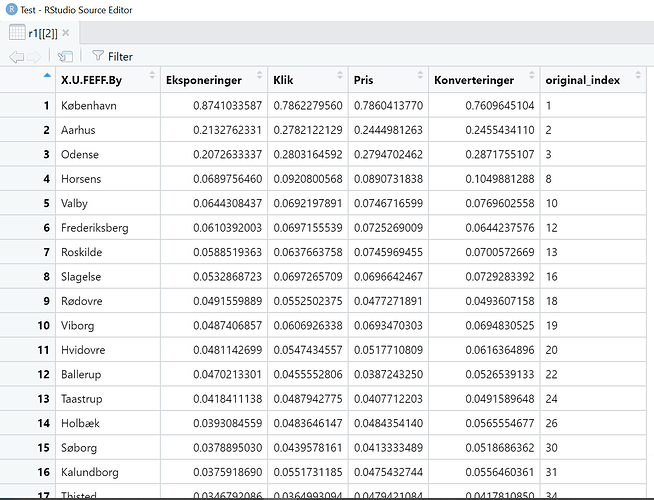
As your can see there is an output for 'global means'. Now it fails at 'r1' object.
Getting closer! However, I am truly clueless as of why. Tried googling the error (and the is_grouped_df function) but with no luck 
mdat <- read.csv2("~/mdat.csv", encoding="UTF-8")
library(tidyverse)
set.seed(42)
normvar <- function(x) {
x / sqrt(sum(x^2))
}
mdat_norm <- mdat %>%
mutate_if(is.numeric, normvar) %>%
mutate(original_index = factor(row_number()))
(global_means <- summarise_if(mdat_norm, .predicate = is.numeric, mean))
#> Eksponeringer Klik Pris Konverteringer
#> 1 0.01743945 0.02045284 0.02129543 0.02212743
cols <- ncol(global_means)
names(global_means) <- paste0("g", names(global_means))
rand_split_2 <- function(x) {
group_map(
.tbl = x %>% mutate(randindex = sample.int(nrow(x), size = nrow(x), replace = FALSE) %% 2) %>% group_by(randindex),
.f = ~ tibble(.)
)
}
r1 <- rand_split_2(mdat_norm)
#> Error in is_grouped_df(.data): argument ".data" is missing, with no default
summarise_a_split <- function(x) {
group_means <- map_dfr(
x,
~ summarise_if(., .predicate = is.numeric, mean)
)
comb <- expand_grid(group_means, global_means)
# rmse
column_rmse <- map_dfc(
names(group_means),
~ (comb[[.]] - comb[[paste0("g", .)]])^2
) %>%
summarise_all(mean) %>%
summarise_all(sqrt)
## sum up the column rmse to get a single num per the config
reduce(column_rmse, .f = `+`)
}
summarise_a_split(r1)
#> Error in map(.x, .f, ...): objekt 'r1' blev ikke fundet
splits_to_do <- 5000
# do x many splits and find the most balanced
splits_x <- map(
1:splits_to_do,
~ rand_split_2(mdat_norm)
)
#> Error in is_grouped_df(.data): argument ".data" is missing, with no default
# summarise them
summaries_x <- map_dbl(
splits_x,
~ summarise_a_split(.)
)
#> Error in map_dbl(splits_x, ~summarise_a_split(.)): objekt 'splits_x' blev ikke fundet
# find the best (norm)
(best_fit <- which(summaries_x == min(summaries_x)))
#> Error in which(summaries_x == min(summaries_x)): objekt 'summaries_x' blev ikke fundet
summaries_x[(best_fit - 2):(best_fit + 2)]
#> Error in eval(expr, envir, enclos): objekt 'summaries_x' blev ikke fundet
best <- splits_x[[best_fit]]
#> Error in eval(expr, envir, enclos): objekt 'splits_x' blev ikke fundet
# find the worst (norm)
(worst_fit <- which(summaries_x == max(summaries_x)))
#> Error in which(summaries_x == max(summaries_x)): objekt 'summaries_x' blev ikke fundet
summaries_x[worst_fit]
#> Error in eval(expr, envir, enclos): objekt 'summaries_x' blev ikke fundet
worst <- splits_x[[worst_fit]]
#> Error in eval(expr, envir, enclos): objekt 'splits_x' blev ikke fundet
# (norm)
map_dfr(
best,
~ summarise_if(., .predicate = is.numeric, mean)
)
#> Error in map(.x, .f, ...): objekt 'best' blev ikke fundet
map_dfr(
worst,
~ summarise_if(., .predicate = is.numeric, mean)
)
#> Error in map(.x, .f, ...): objekt 'worst' blev ikke fundet
walk(
best,
~ print(table(pull(., X.U.FEFF.By)))
)
#> Error in map(.x, .f, ...): objekt 'best' blev ikke fundet
walk(
worst,
~ print(table(pull(., X.U.FEFF.By)))
)
#> Error in map(.x, .f, ...): objekt 'worst' blev ikke fundet
indexes_to_use <- map(
best,
~ pull(., original_index) %>% as.integer()
) %>%
enframe(value = "original_index", name = "group") %>%
unnest(cols = c(original_index)) %>%
arrange(original_index)
#> Error in map(best, ~pull(., original_index) %>% as.integer()): objekt 'best' blev ikke fundet
new_mdat <- bind_cols(indexes_to_use, mdat)
#> Error in list2(...): objekt 'indexes_to_use' blev ikke fundet
new_mdat_split <- group_by(new_mdat, group) %>% group_map(~ tibble(.))
#> Error in group_by(new_mdat, group): objekt 'new_mdat' blev ikke fundet
Created on 2020-07-31 by the reprex package (v0.3.0)