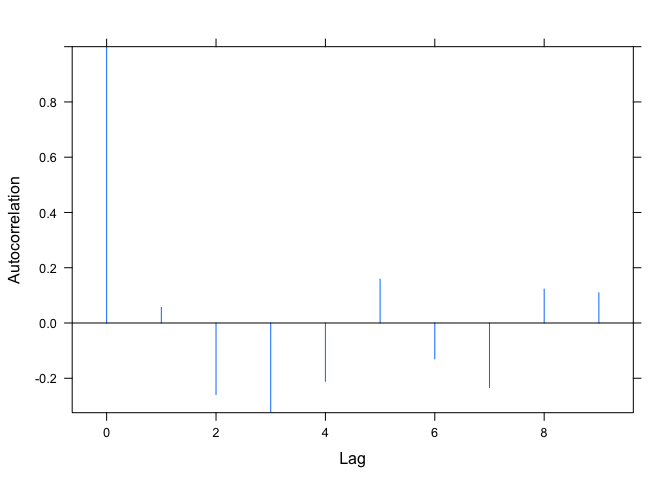
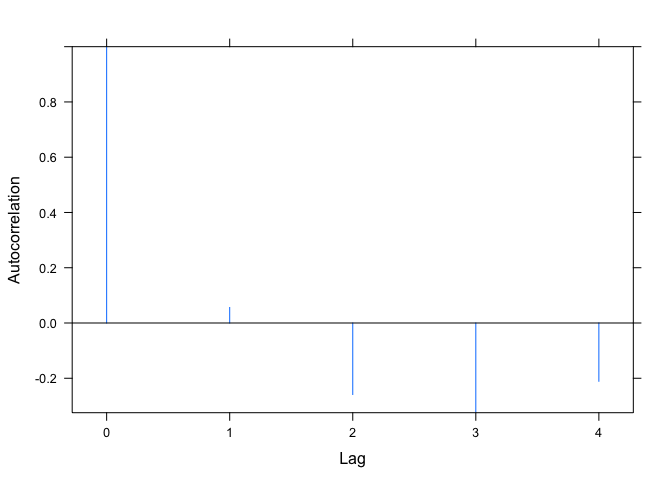
Your second question is a bit more involved (if we continue to use ACF to get the autocorrelation function values), because the plot.ACF function hard-codes the y-axis limits to be the range of the autocorrelations. More on that in a moment. First, an easy way around this: In my example above, m1 is the model. We can directly extract the residuals with resid(m1). Then we can use the base acf function to plot their autocorrelation function. The acf function allows us to explicitly set the y-axis range, rather than hard-coding it inside the function.
acf(resid(m1), lag.max=4, ylim=c(-1,1))
If you still want to use the ACF function, we need to modify the function, plot.ACF that plots ACF objects. To see the function that plots ACF objects, run getAnywhere(plot.ACF) and the code for this function will appear in the console (see the explanatory note at the bottom for some background on why we're looking for the plot.ACF function when we actually called the plot function in our code).
Note that the second line of the function body is ylim <- range(object$ACF). A way around this is to modify the function to accept a ylim argument. Note in the code below that I've created a new function called my_plot.ACF. I've commented out the ylim line in the original function and added a new function argument ylim=c(-1.0, 1.0), so it defaults to full range, but can be changed, if desired, when calling the function.
my_plot.ACF = function (x, alpha = 0, xlab = "Lag", ylab = "Autocorrelation",
grid = FALSE, ylim=c(-1.0,1.0), ...) {
object <- x
#ylim <- range(object$ACF)
if (alpha) {
assign("stdv", qnorm(1 - alpha/2)/sqrt(attr(object, "n.used")))
stMax <- max(stdv)
ylim <- c(min(c(-stMax, ylim[1])), max(c(ylim[2], stMax)))
}
assign("alpha", as.logical(alpha))
assign("grid", grid)
xyplot(ACF ~ lag, object, ylim = ylim, panel = function(x,
y, ...) {
x <- as.numeric(x)
y <- as.numeric(y)
if (grid)
panel.grid()
panel.xyplot(x, y, type = "h")
panel.abline(0, 0)
if (alpha) {
llines(x, stdv, lty = 2)
llines(x, -stdv, lty = 2)
}
}, xlab = xlab, ylab = ylab, ...)
}
Now we run the function. We also need to load the lattice package, since plot.ACF (and therefore our new function) use the xyplot function from lattice.
library(lattice)
my_plot.ACF(ACF(m1, maxLag=4))
Explanatory note in case you're not already familiar with generic functions, methods, and method dispatch: plot is a "generic" function. When you run the plot function, what it does depends on the type of object you pass to it. If you type plot(mtcars) you'll get a scatterplot matrix. If you type plot(lm(mpg ~ hp, data=mtcars)) you'll get several diagnostic plots for a linear regression model. If you type plot(ACF(m1)) you'll get a plot of an autocorrelation function. And so on.
plot does different things with each of these objects because each of them has a different class, specifically, "data.frame", "lm", and "ACF", respectively (for example, run class(ACF(m1)). plot itself just "dispatches" the appropriate "plot method" (the actual function that does the plotting) for each of these objects. To see what these methods are, run methods(plot). They'll all be called plot.zzz where zzz is the class of the object.. For example, note that one of the functions is plot.ACF. You can see the code for this function by typing getAnywhere(plot.ACF).