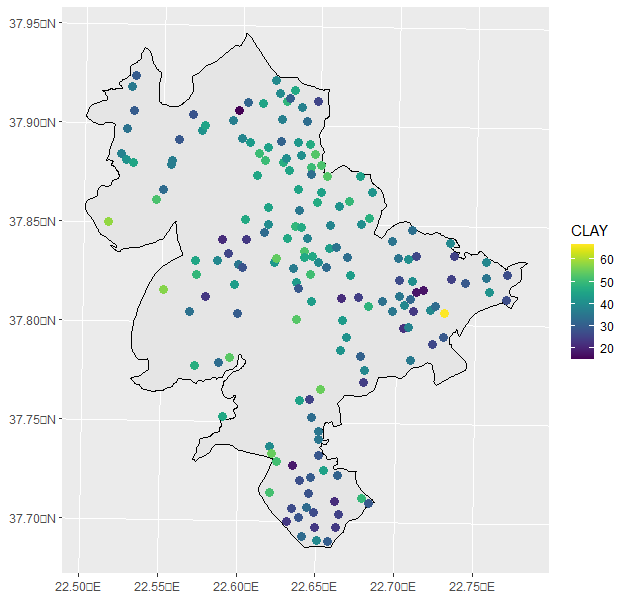
I have run my data in R, and created a distribution mapof my data (Clay Soil Property).
I want to detect in a way the spatial outliers that i can see from the map, and remove them in order to execute kriging NOT in all the set, but in the remaining data (except the outliers).
Do you have any idea with the code i should write?
Until now, i wrote the following code for the creation of the distribution map:
#Next steps for Kriging Methods
boundary <- st_read("Aoi.shp") mycrs <- st_crs(boundary)
mydata <- read.csv("Attributesfixed.csv", sep = ";") mydata2 <- mydata %>% select("X","Y","PH","CACO3","SAND","SILT", "CLAY" ,"OM","CEC")
mydata2 <- st_as_sf(mydata2, coords=c("X","Y"), crs=mycrs) ggplot() + geom_sf(data=boundary, color="black", size=1) + geom_sf(data=mydata2, aes(color=CLAY), size=3) + scale_color_viridis() And i plotted the map you can see:
This is the code i wrote for the ordinary kriging BUT in the whole set. #Mask for ordinary kriging
mask <- read_stars("AoiGrid") st_crs(mask) <- mycrs plot(mask)
ordinary kriging
mydata2.krig <- krige(CLAY~1, mydata2, newdata=mask, vgm)
names(mydata2.krig) names(mydata2.krig)1 <- "CLAY.pred" names(mydata2.krig)
min(mydata2.krig$CLAY.pred, na.rm=T); max(mydata2.krig$CLAY.pred, na.rm=T)
ggplot() + geom_stars(data=mydata2.krig["CLAY.pred"]) + scale_fill_gradient(low="yellow", high="dark blue", limits=c(18,60)) + geom_sf(data=mydata2, shape=1, aes(size=CLAY))
I want to run it in the set without the outliers, that i will previously remove in some way! Any help?
Thanks in advance!