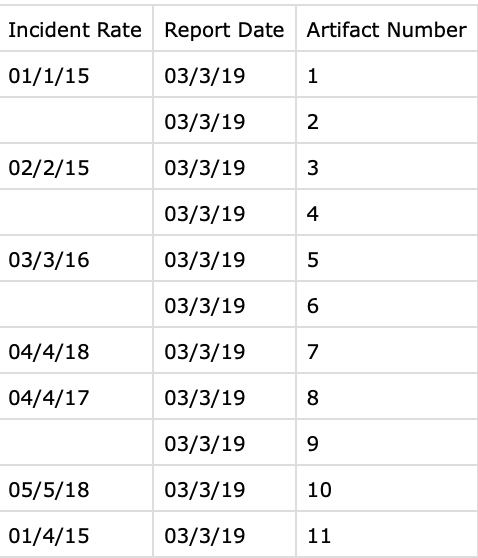
Yes, it helps, this is sample data on a proper format
You are getting 'NA' as a string (with quotes) because you are not reading the data correctly from the CSV file, they should be NA (without quotes) which is the way R represents blanks, it stands for Not Available.
Anyways, If I understand you correctly, this is what you are trying to do
library(tidyverse)
library(lubridate)
# This is just sample data, you can replace this with the actual dataset that you read from the CSV file
sample_data <- data.frame(
Incident.Date = c('1/1/2015','NA','2/2/15','NA','5/5/2016','NA','4/4/2017','4/4/2018','NA','5/5/2018','1/4/2015'),
Report.Date = c('3/3/2019','3/3/2019','3/3/2019','3/3/2019','3/3/2019','3/3/2019','3/3/2019','3/3/2019','3/3/2019','3/3/2019','3/3/2019'),
Artifact.Number = c(1,2,3,4,5,6,7,8,9,10,11)
)
sample_data %>%
mutate_at(vars(contains("Date")), dmy) %>%
rowwise() %>%
mutate(Incident.Date = if_else(
is.na(Incident.Date),
true = Report.Date,
false = Incident.Date)
) %>%
ungroup() %>%
mutate(year = year(Incident.Date),
month = month(Incident.Date)) %>%
arrange(Incident.Date)
#> Warning: 4 failed to parse.
#> # A tibble: 11 x 5
#> Incident.Date Report.Date Artifact.Number year month
#> <date> <date> <dbl> <dbl> <dbl>
#> 1 2015-01-01 2019-03-03 1 2015 1
#> 2 2015-02-02 2019-03-03 3 2015 2
#> 3 2015-04-01 2019-03-03 11 2015 4
#> 4 2016-05-05 2019-03-03 5 2016 5
#> 5 2017-04-04 2019-03-03 7 2017 4
#> 6 2018-04-04 2019-03-03 8 2018 4
#> 7 2018-05-05 2019-03-03 10 2018 5
#> 8 2019-03-03 2019-03-03 2 2019 3
#> 9 2019-03-03 2019-03-03 4 2019 3
#> 10 2019-03-03 2019-03-03 6 2019 3
#> 11 2019-03-03 2019-03-03 9 2019 3
Created on 2020-01-20 by the reprex package (v0.3.0.9000)