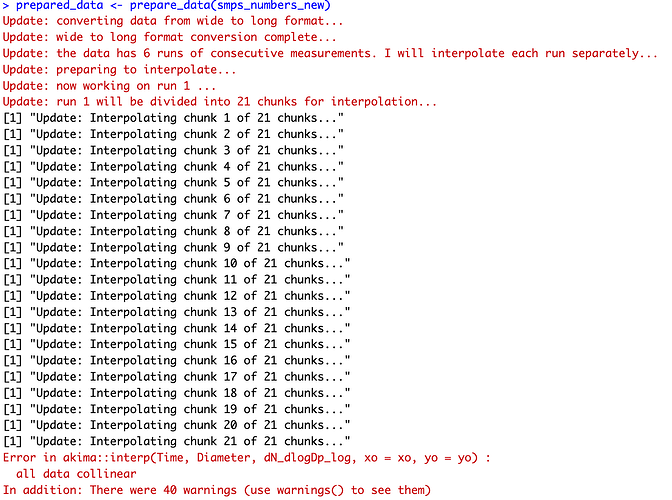
I do not see that. Your data has 19359 rows of nothing but NA's. So out of 28535188 data points 2071413 of them are NA.
You really cannot see this in R or a spreadsheet but if you open the .csv file in a text editor this is what the last two rows of your raw data look like
,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
There in 106 commas in a row. And you have 19359 of these rows.
There are no NA's in the example data.
My guess is that you have an instrument malfunction in whatever machine is measuring those particle sizes.