Hi all,
I'm reading in a very small .csv file to get some simple trajectory plots. There look are
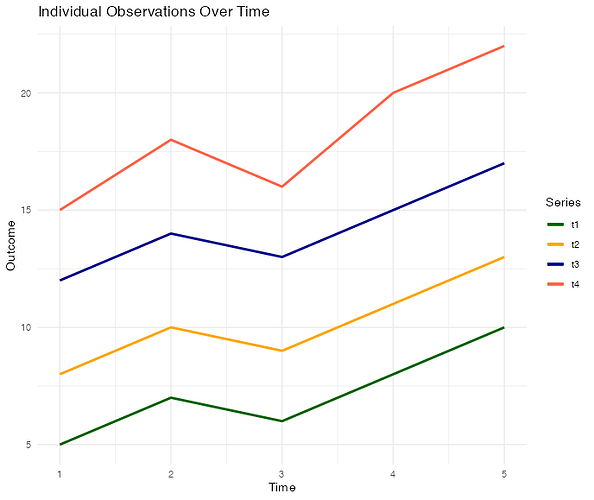
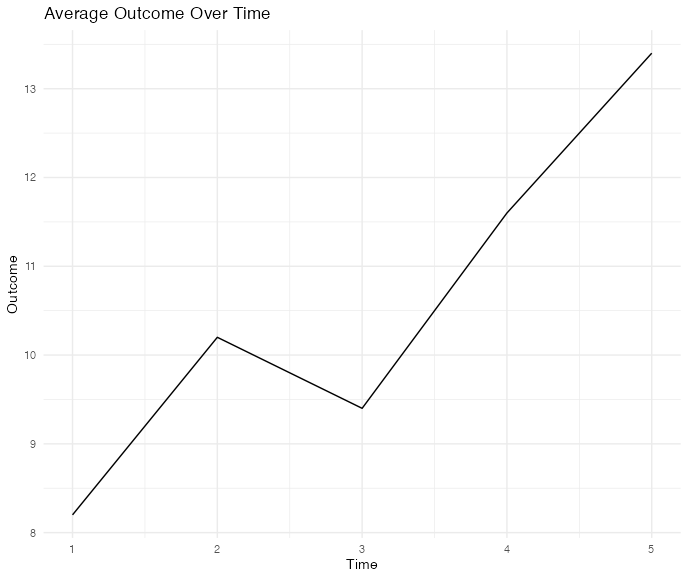
t1 t2 t3 t4
5 8 12 15
7 10 14 18
6 9 13 16
8 11 15 20
10 13 17 22
The app I'm writing so far reads in the data and I simply want ggplot to give me a plot of the average value across these individuals with an overlay of the individual lines, in another color. The code chunk is
observeEvent(input$run_button, {
print("Button Clicked")
req(input$outcome_variable, input$predictor_variable, input$time_indicator, input$data_file)
df <- read.csv(input$data_file$datapath)
outcome_var <- input$outcome_variable
# Simplified example of creating a line plot for average outcome over time
average_plot <- ggplot(df, aes(x = 1:ncol(df), y = df[, outcome_var])) +
geom_line() +
labs(title = "Average Outcome Over Time", x = "Time", y = "Outcome")
# Simplified example of creating line plots for individual observations over time
individual_plot <- ggplot(df, aes(x = 1:ncol(df), y = df[, outcome_var])) +
geom_line(aes(group = 1:nrow(df), color = as.factor(1:nrow(df)))) +
labs(title = "Individual Observations Over Time", x = "Time", y = "Outcome") +
theme_minimal()
# Render the plots without using print
output$average_plot <- renderPlot({
average_plot
})
output$individual_plot <- renderPlot({
individual_plot
})
})
}
# Run the Shiny app
shinyApp(ui, server)
When a click the button called "Create plots", nothing shows up, but there is no error either. The
print("Button Clicked")
line was just so I could tell if it saw the Create plots button. It did. Your suggestions are welcomed.
Thank you
D