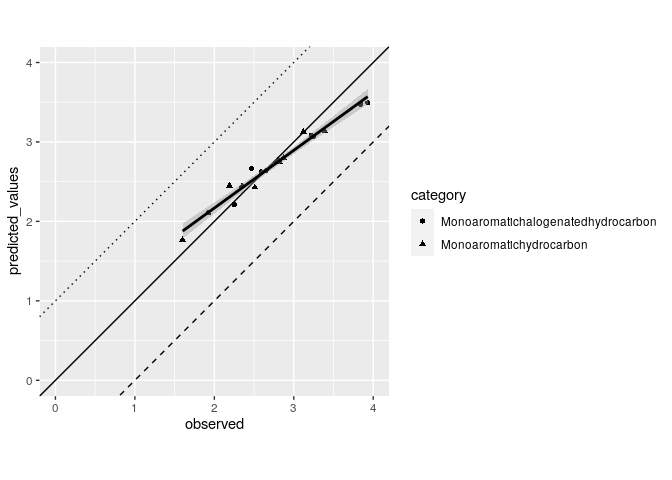
This can work as a starting point
library(tidyverse)
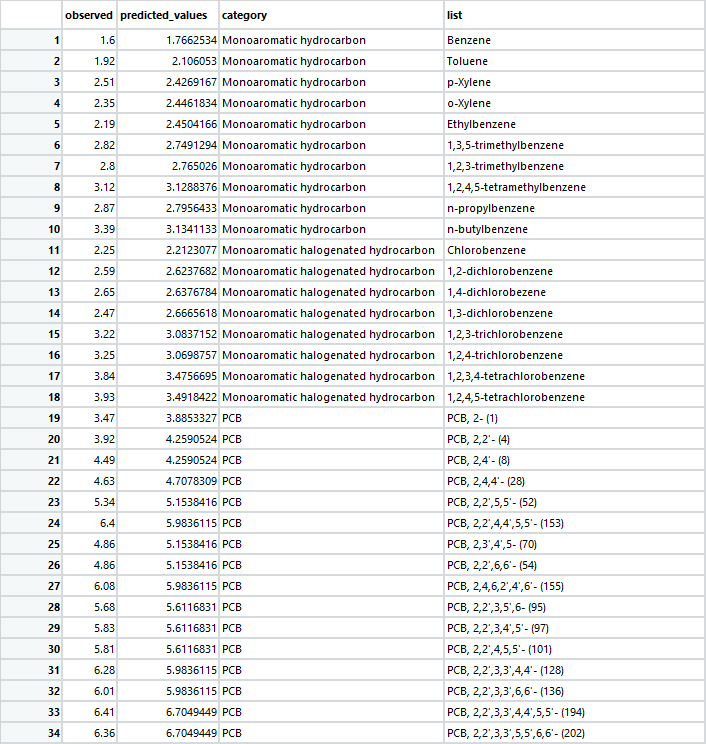
testdata1 = tibble::tribble(
~observed, ~predicted_values, ~category, ~list,
1.6, 1.7662534, "Monoaromatichydrocarbon", "Benzene",
1.92, 2.106053, "Monoaromatichydrocarbon", "Toluene",
2.51, 2.4269167, "Monoaromatichydrocarbon", "p-Xylene",
2.35, 2.4461834, "Monoaromatichydrocarbon", "o-Xylene",
2.19, 2.4504166, "Monoaromatichydrocarbon", "Ethylbenzene",
2.82, 2.7491294, "Monoaromatichydrocarbon", "1,3,5-trimethylbenzene",
2.8, 2.765026, "Monoaromatichydrocarbon", "1,2,3-trimethylbenzene",
3.12, 3.1288376, "Monoaromatichydrocarbon", "1,2,4,5-tetramethylbenzene",
2.87, 2.7956433, "Monoaromatichydrocarbon", "n-propylbenzene",
3.39, 3.1341133, "Monoaromatichydrocarbon", "n-butylbenzene",
2.25, 2.2123077, "Monoaromatichalogenatedhydrocarbon", "Chlorobenzene",
2.59, 2.6237682, "Monoaromatichalogenatedhydrocarbon", "1,2-dichlorobenzene",
2.65, 2.6376784, "Monoaromatichalogenatedhydrocarbon", "1,4-dichlorobezene",
2.47, 2.6665618, "Monoaromatichalogenatedhydrocarbon", "1,3-dichlorobenzene",
3.22, 3.0837152, "Monoaromatichalogenatedhydrocarbon", "1,2,3-trichlorobenzene",
3.25, 3.0698757, "Monoaromatichalogenatedhydrocarbon", "1,2,4-trichlorobenzene",
3.84, 3.4756695, "Monoaromatichalogenatedhydrocarbon", "1,2,3,4-tetrachlorobenzene",
3.93, 3.4918422, "Monoaromatichalogenatedhydrocarbon", "1,2,4,5-tetrachlorobenzene"
)
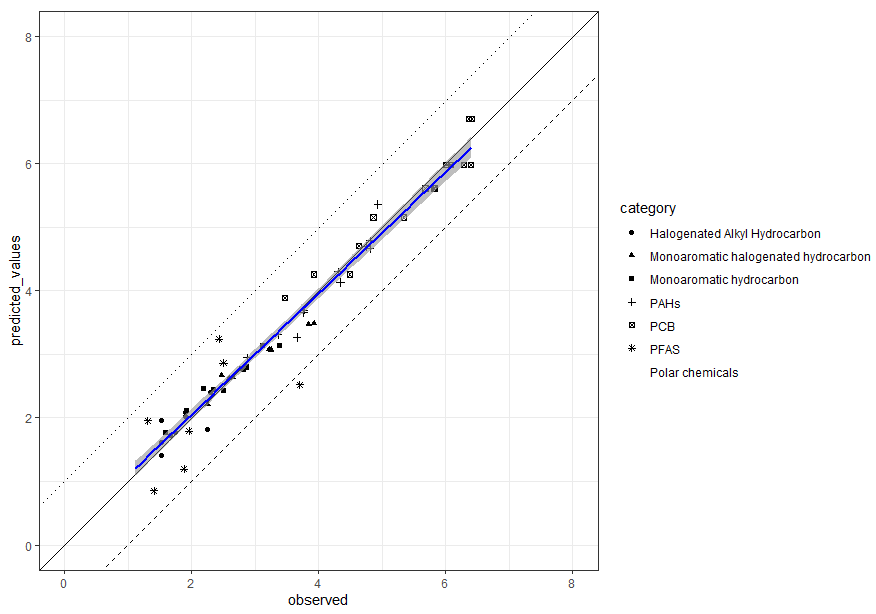
testdata1 %>%
ggplot(aes(x = observed, y = predicted_values)) +
geom_point(aes(shape = category)) +
geom_abline(slope = 1, intercept = 0) +
geom_abline(slope = 1, intercept = 1,
linetype = "dotted") +
geom_abline(slope = 1, intercept = - 1,
linetype = "dashed") +
scale_x_continuous(limits = c(0, 4)) +
scale_y_continuous(limits = c(0, 4)) +
geom_smooth(method = "lm", color = "black") +
coord_equal()
#> `geom_smooth()` using formula 'y ~ x'