Hallo R studio Community.
I am having an issue with the interpration of glm function.
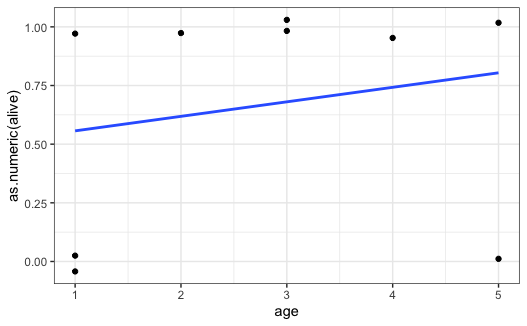
Let's consider this df
df = data.frame(alive = c("FALSE","TRUE","FALSE","FALSE","TRUE","TRUE","TRUE","TRUE","TRUE"),
sex = rep(c("M","M","F"),3),
age =c(1,1,1,5,3,2,5,4,3))
and then I reassess variables classes
df$sex <- as.factor(df$sex)
df$alive <- as.logical(df$alive)
Now I fit my regression
f1 <- glm(alive ~ age, data = df, family = "binomial")
summary(f1)
tidy(f1, exponentiate = TRUE, conf.int = TRUE)
here I get
> tidy(f1, exponentiate = TRUE, conf.int = TRUE)
# A tibble: 2 x 7
term estimate std.error statistic p.value conf.low conf.high
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 0.924 1.42 -0.0556 0.956 0.0482 19.1
2 age 1.34 0.484 0.600 0.548 0.535 4.12```
My question: should the dicotomic outcome (alive) be expressed in logical class or in numeric class ( 0 / 1)? And if it is numeric class, should be "no outcome" = 0 and
"yes outcome"= 1 ?
I am having trouble with my real life dataset, because I get strange odds ratio ( the opposite for what I expected, and I am afraid I am missing something.
Compared to the example in the reprex, in my real life dataset is like if I would have <1 estimated odds ratio, even if I am sure that increasing age is a risk factor for mortality.
Thanks in advance for your help!