I have data with different variables:
The code:
pm_hourly %>%
select(date,HOUSE_SO4,HOUSE_ratio_SO4_OA,HOUSE_NOx_NEW,HOUSE_NO2_ppb,`relative_humidity_%`) %>%
melt(id.vars=1:1) -> HOURLYa
levels(HOURLYa$variable) <- c( "AMS_SO[4]",
"AMS(SO[4]:OA)",
"NO[x]~ (ppb)",
"NO[2]~ (ppb)",
"RH(`%`)")
AMS_SO4_NOX_SO2_HOUSE_HOURLYaa %>%
filter(date >= as.Date("2022-01-27 00:00") & date <= as.Date("2022-02-06 23:59")) %>%
ggplot(aes(x = date, y = value, color = variable, fill = variable, group = variable)) +
geom_area(data = . %>% filter(variable != "RH(`%`)", variable != "NO[x]~ (ppb)"), alpha = 0.6) +
geom_line(data = . %>% filter(variable == "RH(`%`)"), size = 1.5) +
geom_line(data = . %>% filter(variable == "NO[x]~ (ppb)"), size = 1.5, linetype = "dashed") +
xlab("(2022) Hourly") +
ylab("") +
labs(title = "HOUSE Site") +
theme(legend.position = "right") +
theme(axis.title = element_text(face = "plain", size = 16, color = "black"),
axis.text = element_text(size = 16, face = "plain", color = "black"),
axis.title.x = element_text(vjust = 0.1),
axis.text.y = element_text(hjust = 0.5),
plot.title = element_text(size = 15)) +
theme(strip.text = element_text(size = 12, color = "black")) +
scale_x_datetime(expand = c(0, 0),
date_breaks = "2 days",
date_minor_breaks = "5 days",
date_labels = "%m/%d",
limits = as.POSIXct(c("2022-01-26 00:00:00", "2022-02-05 20:59:00"))) +
scale_y_continuous(sec.axis = sec_axis(~., name = "AMS(SO[4]:OA)",
breaks = seq(0, 1, by = 0.1),
labels = seq(0, 1, by = 0.1)))
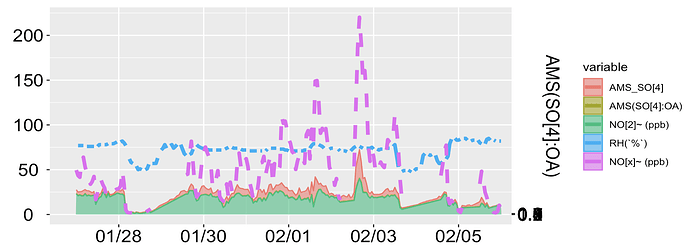
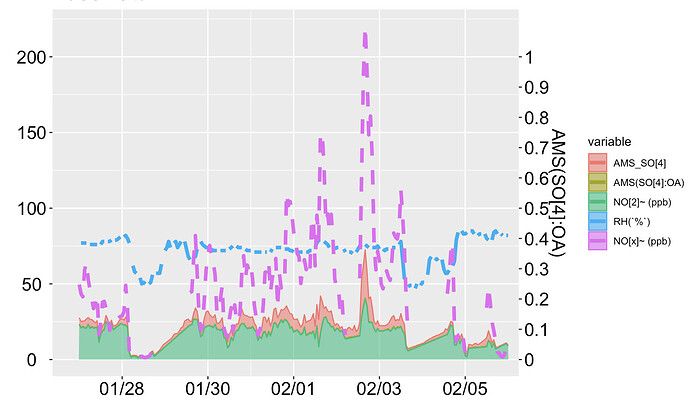
Now, I want to shift "AMS(SO[4]:OA)" to the right secondary axis with the limit 0 to 1.
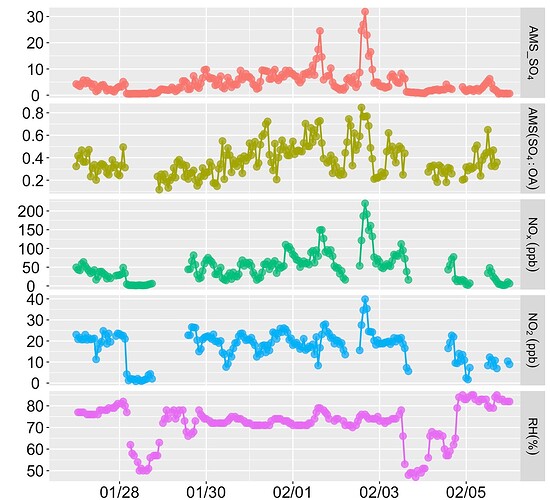
Also, I don't want a stacked filled area plot, because, AMS_SO[4] has lower values than NO[2] and from this (filled area) plot, it looks like AMS_SO[4] has a higher value than NO[2]. I have also attached the line plot.
Thanks