I am trying to read in a bunch of text files , get them into tidy format and arrange them into bigrams first having got rid of stop_words and a bunch of words I saved in a df called rare_words. It all works fine until after unnest_tokens I come to separating into first and second word, so as to anti_join on stop_words and rare_words. At this point something happens, which I don't understand: I am getting an extra column with t's and s's etc where words like "sherif's department" is split into "sherif" , "s" and "department"; otherwise, if a bigram doesn't contain an apostrophe, the field shows NA.
I also get this message:
Warning messages:
1: Expected 3 pieces. Additional pieces discarded in 39161 rows [34515, 35383, 35384, 35385, 35386, 35388, 65758, 68458, 73653, 86848, 92074, 108182, 129475, 138778, 139845, 149475, 167656, 186861, 206700, 217459, ...].
2: Expected 3 pieces. Missing pieces filled with `NA` in 11140778 rows [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, ...].
Googling this message hasn't really given me any answers.
The code is bellow:
txt_files <- list.files(pattern = ".txt")
bigrams <- list.files(pattern = "*.txt") %>%
map_chr(~ read_file(.)) %>%
tibble(text = .) %>%
drop_na() %>%
mutate(filename = txt_files) %>%
unnest_tokens(word, text, token = 'ngrams', n = 2) %>%
count(word, sort = TRUE) %>%
# Here's where the problem appears
separate(word, into =c('first_word', 'second_word', sep=' ') ) %>%
anti_join(stop_words, by=c(first_word='word' ) ) %>%
anti_join(stop_words, by=c(second_word='word' ) ) %>%
anti_join(rare_words, by=c(first_word='word') ) %>%
anti_join(rare_words, by=c(second_word= 'word')) %>%
mutate(ngram = paste(first_word, second_word))
And I end up with the following:
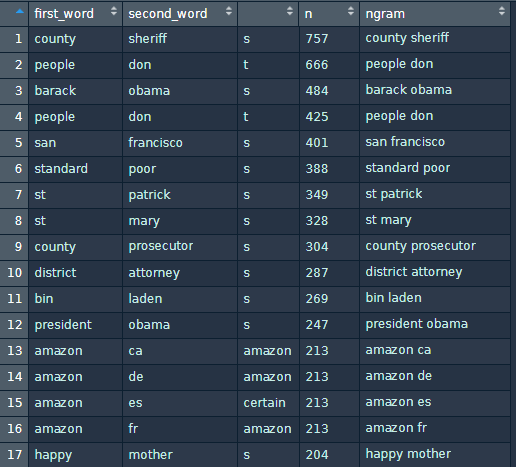
I would really appreciate if someone could explain how I can work around this problem, I mean how can I create a df with just the first word, second word, count and bigram, without splitting words with apostrophes. Thank you.