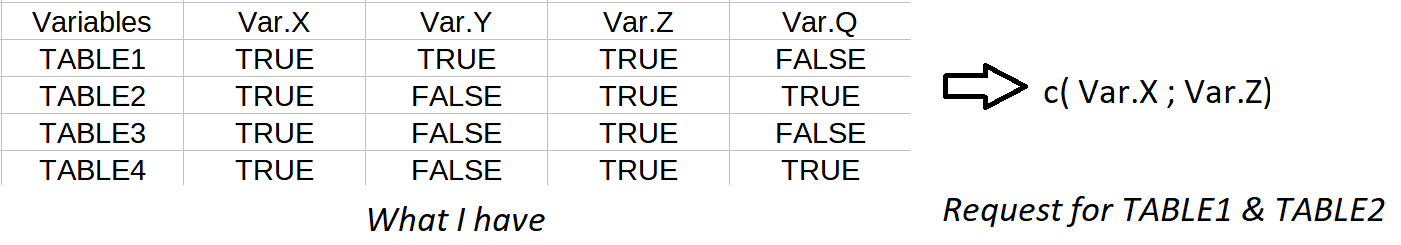
Maybe like this?
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
df1 <- tibble::tribble(
~table,~Var.X,~Var.Y,~Var.Z,~Var.Q,
"TABLE1",T,T,T,F,
"TABLE2",T,F,T,T,
"TABLE3",T,F,T,F,
"TABLE4",T,F,T,T
)
varnames <- setdiff (names(df1),"table")
# combine all rows to compare table variables
df1 <- df1 |>
mutate(seqnr=row_number())
df1a <-df1
names(df1a) <- paste0("a",names(df1a))
df1b <-df1
names(df1b) <- paste0("b",names(df1b))
df2 <- tidyr::crossing(df1a,df1b) |>
filter(aseqnr>bseqnr) |>
select(-c(aseqnr,bseqnr)) |>
rename(table1=atable,table2=btable)
# variable exists in two tables
anames <- paste0("a",varnames)
bnames <- paste0("b",varnames)
df3 <- cbind(df2[,c("table1","table2")],(df2[,anames]==T) & (df2[,bnames]==T))
names(df3) <- c("table1","table2",varnames)
# copy variable name
df4 <- df3 |>
mutate(across(
.cols=!any_of(c("table1","table2")),
.fns = \(x) {cc <-c("",cur_column())[1+x] ; return(cc)}
))
# combine variable names in column m
df4 |>
rowwise() |>
mutate(m=paste(c_across(!any_of(c("table1","table2"))),collapse = " ")) |>
select(table1,table2,m) |>
mutate(m=stringr::str_squish(m)) |>
print()
#> # A tibble: 6 × 3
#> # Rowwise:
#> table1 table2 m
#> <chr> <chr> <chr>
#> 1 TABLE2 TABLE1 Var.X Var.Z
#> 2 TABLE3 TABLE1 Var.X Var.Z
#> 3 TABLE3 TABLE2 Var.X Var.Z
#> 4 TABLE4 TABLE1 Var.X Var.Z
#> 5 TABLE4 TABLE2 Var.X Var.Z Var.Q
#> 6 TABLE4 TABLE3 Var.X Var.Z