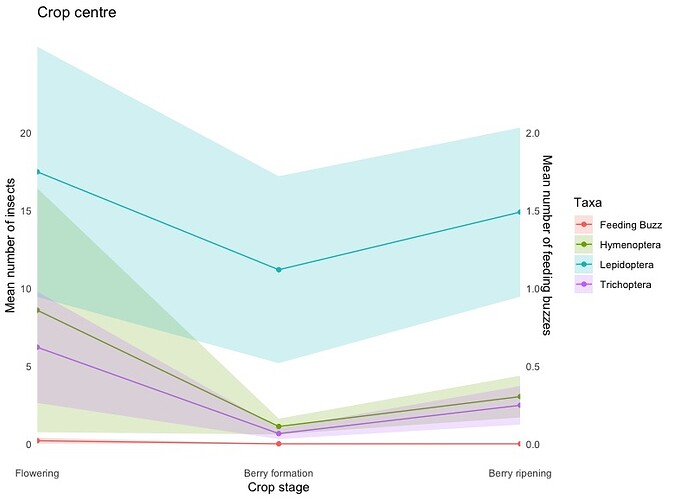
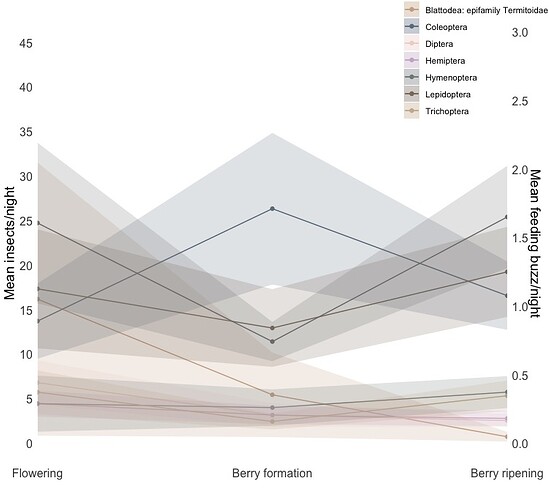
Hi,
I'm very much a beginner user and have gotten my code to this point but I don't seem to be able to work out how to link the feeding buzz data to the secondary y-axis, it just seems to follow the primary axis values. All the data appears to be correct except for the secondary y-axis.
I appreciate the help, I'm lost.
2023-12-14T14:00:00Z
# Your data
data <- data.frame(
Stage = rep(c("Flowering", "Berry formation", "Berry ripening"), each = 4),
Taxa = rep(c("Hymenoptera", "Lepidoptera", "Trichoptera", "Feeding Buzz"), times = 3),
Value = c(
8.595238095, 17.5, 6.214285714, 0.2,
1.114285714, 11.2, 0.657142857, 0,
3.038961039, 14.90909091, 2.480519481, 0))
# Standard error data
error_data <- data.frame(
Stage = rep(c("Flowering", "Berry formation", "Berry ripening"), each = 4),
Taxa = rep(c("Hymenoptera", "Lepidoptera", "Trichoptera", "Feeding Buzz"), times = 3),
SE = c(
7.846380726, 8.053050727, 3.582532101, 0.2,
0.50836933, 6.012843397, 0.354900524, 0,
1.344521498, 5.438407432, 1.238978808, 0))
# Merge data and error_data
merged_data <- merge(data, error_data, by = c("Stage", "Taxa"))
# Reorder the stages
merged_data$Stage <- factor(merged_data$Stage, levels = c("Flowering", "Berry formation", "Berry ripening"))
# Create ggplot for a line graph with shaded error bands using geom_ribbon
ggplot(merged_data, aes(x = Stage, group = Taxa)) +
geom_line(aes(y = Value, color = Taxa)) +
geom_point(aes(y = Value, color = Taxa)) +
geom_ribbon(aes(ymin = Value - SE, ymax = Value + SE, fill = Taxa), alpha = 0.2) +
scale_y_continuous(
name = "Mean number of insects",
breaks = seq(0, 20, 5),
sec.axis = sec_axis(~. *.1, name = "Mean number of feeding buzzes", breaks = seq(0, 2, 0.5))) +
scale_x_discrete(expand = c(0, 0)) +
labs(title = "Crop centre",
x = "Crop stage") +
theme_minimal() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank())