snt
August 13, 2018, 3:15pm
1
I have an output to be rendered in a shiny app using DT::datatable.
The output which currently looks like this and I want to use the second row as the column headers:
tab1:
V1 V2 V3 V4 V5 V6 V7 V8
Month Apr-17 May-17 Jun-17 Jul-17 Aug-17 Sep-17 Oct-17 Nov-17
aaa 116.719 120.404 120.26 123.431 117.327 110.742 114.811 117.34
bbb 76.118 75.976 76.215 76.134 77.19 78.519 78.258 74.522
So when I want to use the row month as column headers I do the following:
app1 <-reactive({ tab1()%>% .[-1,]})
And then I render it using datatable
output$op1 <-renderDataTable({
app1()
})
I get the following output :
V1 V2 V3 V4 V5 V6 V7 V8
aaa 116.719 120.404 120.26 123.431 117.327 110.742 114.811 117.34
bbb 76.118 75.976 76.215 76.134 77.19 78.519 78.258 74.522
How do I replace the v1,v2 by month names .Thank you.
cderv
August 13, 2018, 3:26pm
2
I think you should prepare you data.frame before DT::datatable
Add the first row as names for the DF
delete the first row
With base R
df <- data.frame(V1 = c("Letter", "A", "B"), V2 = c("Number", "1", "2"), stringsAsFactors = FALSE)
names(df) <- df[1, ]
df <- df[-1, ]
df
#> Letter Number
#> 2 A 1
#> 3 B 2
Created on 2018-08-13 by the reprex package (v0.2.0).
with tidyverse
library(tidyverse)
df <- tibble(V1 = c("Letter", "A", "B"), V2 = c("Number", "1", "2"))
df %>%
# rename with first row
set_names(df[1, ]) %>%
# delete the first row
slice(-1)
#> # A tibble: 2 x 2
#> Letter Number
#> <chr> <chr>
#> 1 A 1
#> 2 B 2
Created on 2018-08-13 by the reprex package (v0.2.0).
snt
August 13, 2018, 3:47pm
3
I get this error when I do this nm must be NULL or a character vector the same length as x
cderv
August 13, 2018, 4:08pm
4
set_names but I get no error on my side.
library(tidyverse)
df <- set_names(df, nm = df[1, ])
df <- slice(df, -1)
Does it works ?df %>% set_names(df[1, ]) %>% slice(-1)
snt
August 13, 2018, 4:20pm
5
I am pretty much sure the solution you provided is correct. A pre build up to what I am doing to get this point is
intrc_pattern <-reactive({MyData %>%
filter(Tab == "Interaction pattern" & Classification_Level == input$selected_class & Primary_family == input$selected_product &
Metric_name == input$selected_metric ) %>%
mutate(`pct` = as.numeric(as.character(Metric_Value))) %>%
mutate(`Month` = as.yearmon(Month_considered, "%b-%y")) %>%
select(Month,`pct`,Secondary_family)%>%
group_by(Month) %>% na.omit()
})
intrc_pattern_reshape <-reactive({ dcast(intrc_pattern(), Month ~ Secondary_family, value.var="pct", fill=0)%>% t() %>% .[-1,]})
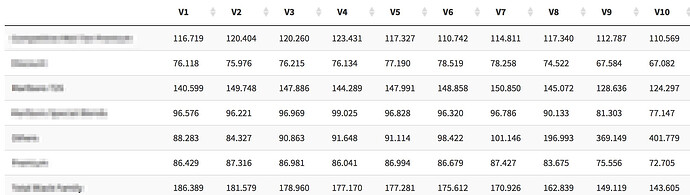
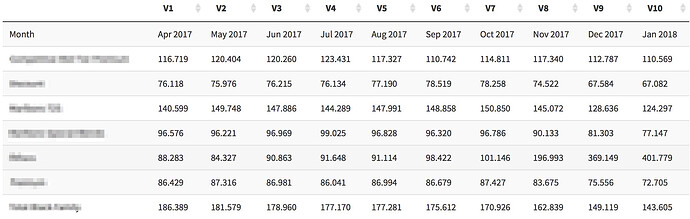
If i do use .[-1.] then my output is like this
And if I dont do it its like this
I am not sure if this is helpful without the data.
 This error comes from
This error comes from