SARS-CoV-2 Pango Lineage Translator
Authors: @sschmutz
website, twitter
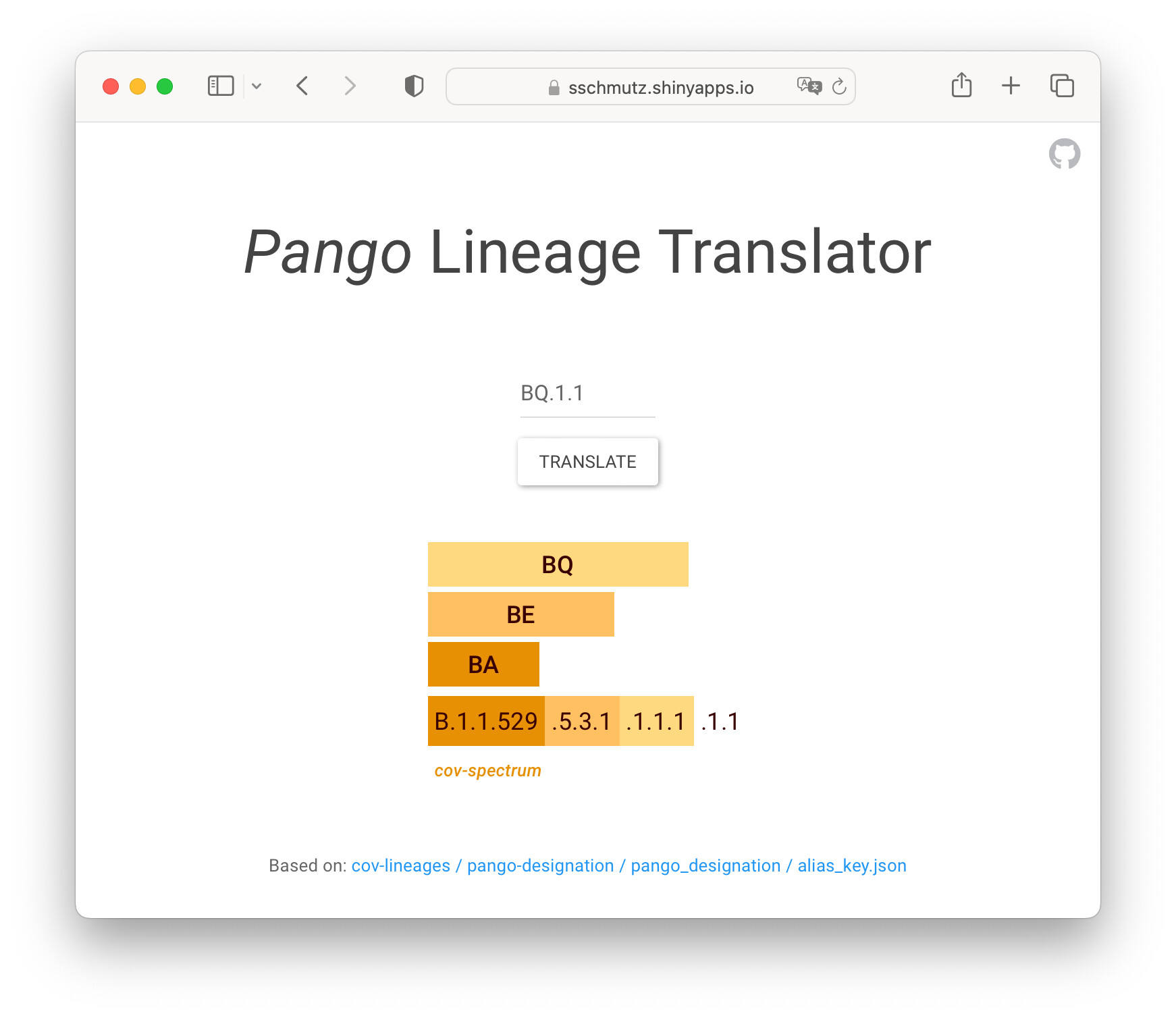
Abstract: The aim of this table, wrapped in an interactive Shiny App, was to help understand the hierarchical structure of SARS-CoV-2 Pango lineage names and their abbreviation.
Full Description: As viruses evolve, new lineages can be designated. The Pango Network introduced a systematic nomenclature system for SARS-CoV-2. There is an alias for each third hierarchical level to simplify these lineage labels. For example, the variant of concern "Omicron" can be described as lineage B.1.1.529.* or shorter, just BA.*.
As evolution continues, lineage labels become longer and longer, and the aliases become more and more nested.
This led Theo Sanderson to compose the following tweet, which inspired me to do this project:

The interactive Shiny App I wrote outputs a color-coded {gt} table that splits these Pango lineages into its parts.
The colors for the nested aliases are lighter variants of a base color (created using gt::adjust_luminance()), which is different for each variant of concern (e.g. Delta = blue, Omicron = orange).
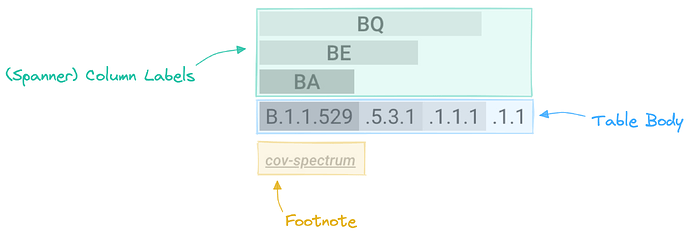
At first glance, it might not be clear how this table is structured. In detail, the column labels listing the aliases (e.g. BA, BE, BQ) consist of spanner column labels, and the complete lineage label is split into its individual parts across multiple cells as part of the table body.
Therefore, for a given lineage, it always returns a table consisting of a single row with as many spanner column labels as there are aliases.
Finally, there is a footnote containing a direct link to the cov-spectrum page of the chosen lineage, where more information on it is shown.
Table Type: interactive-Shiny
Submission Type: Other
Table: https://sschmutz.shinyapps.io/PangoLineageTranslator/
Code: GitHub - sschmutz/PangoLineageTranslator: Translate and Illustrate SARS-CoV-2 Pango Lineages
Cloud project:
Languages: Built with R: true. Built with Python: false.
Industries: Life science.
Other packages: gt