I installed standalone Rstudio version with R.
Separately to this, I have Python environment setup with anaconda.
Python environment works with no issues in jupyter notebook, jupyter lab, spyder IDE or under VScode.
Now I want to use R-markdown for reporting.
In R studio, I start New Project.
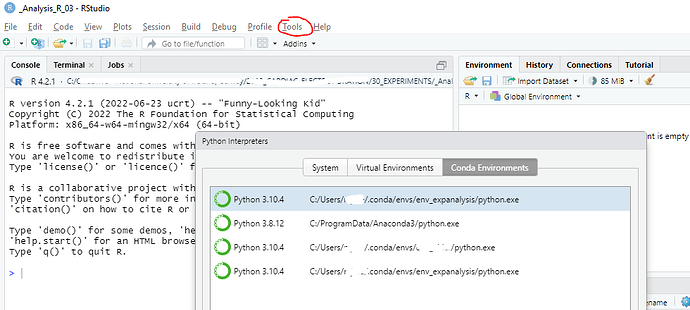
in Tools Menu, Project Options and Global Options, under Python section I choose my conda environment
then I create new R-markdown file in my project directory.
I can knit the template with no issues.
Now I add python chunks:
- environment check:
# check environment
Sys.getenv("RETICULATE_PYTHON")
it returns: [1] "C:/Users/.../.conda/envs/env_expanalysis/python.exe"
also python chunk:
x=42
print(x)
runs with no issues (with both 'run current chunk' and knit).
So, to me it looks like python environment is setup correctly.
I can even plot something with matplotlib running this chunk
import matplotlib.pyplot as plt
import numpy as np
xpoints = np.array([1, 8])
ypoints = np.array([3, 10])
plt.plot(xpoints, ypoints)
plt.show()
This works fine.
Now, I need pandas library for my analysis.. and here is the problem
import pandas as pd
this returns an error:
"ImportError: DLL load failed while importing _bz2: The specified module could not be found."
Of course pandas is installed and works fine in jupyter notebook using the same environment.
Do you have any ideas what is wrong here?
Thanks.