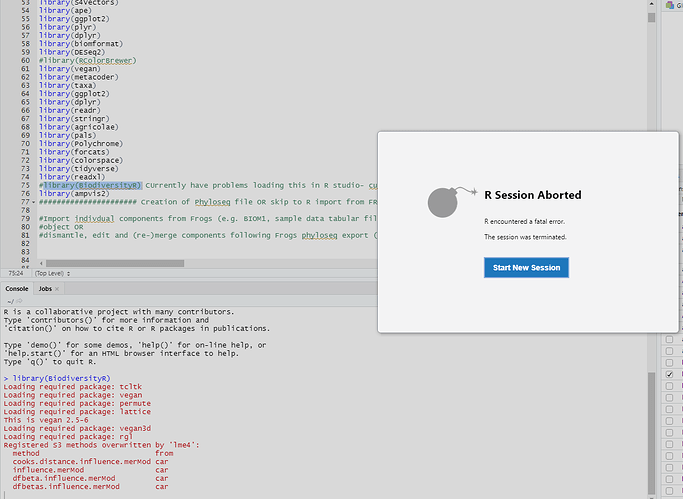
Problem occurs in both the stable (RStudio 1.3.959) and preview versions (RStudio Version 1.3.1023)
sessionInfo()
R version 4.0.1 (2020-06-06)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 17763)
Matrix products: default
locale:
[1] LC_COLLATE=English_United Kingdom.1252 LC_CTYPE=English_United Kingdom.1252 LC_MONETARY=English_United Kingdom.1252
[4] LC_NUMERIC=C LC_TIME=English_United Kingdom.1252
attached base packages:
[1] stats4 parallel stats graphics grDevices utils datasets methods base
other attached packages:
[1] ampvis2_2.6.3 readxl_1.3.1 purrr_0.3.4 tidyr_1.1.0
[5] tibble_3.0.1 tidyverse_1.3.0 colorspace_1.4-1 forcats_0.5.0
[9] Polychrome_1.2.5 pals_1.6 agricolae_1.3-3 stringr_1.4.0
[13] readr_1.3.1 vegan_2.5-6 lattice_0.20-41 permute_0.9-5
[17] DESeq2_1.28.1 SummarizedExperiment_1.18.1 DelayedArray_0.14.0 matrixStats_0.56.0
[21] GenomicRanges_1.40.0 GenomeInfoDb_1.24.0 IRanges_2.22.2 biomformat_1.16.0
[25] dplyr_1.0.0 plyr_1.8.6 ape_5.4 S4Vectors_0.26.1
[29] metagenomeSeq_1.30.0 RColorBrewer_1.1-2 glmnet_4.0-2 Matrix_1.2-18
[33] limma_3.44.3 Biobase_2.48.0 BiocGenerics_0.34.0 ggpubr_0.3.0
[37] ggplot2_3.3.1 DivNet_0.3.6 phyloseq_1.32.0 breakaway_4.6.16
[41] metacoder_0.3.4 taxa_0.3.4
loaded via a namespace (and not attached):
[1] questionr_0.7.1 tidyselect_1.1.0 htmlwidgets_1.5.1 RSQLite_2.2.0 AnnotationDbi_1.50.0
[6] grid_4.0.1 combinat_0.0-8 BiocParallel_1.22.0 devtools_2.3.0 munsell_0.5.0
[11] codetools_0.2-16 miniUI_0.1.1.1 withr_2.2.0 highr_0.8 AlgDesign_1.2.0
[16] rstudioapi_0.11 ggsignif_0.6.0 GenomeInfoDbData_1.2.3 bit64_0.9-7 rhdf5_2.32.0
[21] rprojroot_1.3-2 vctrs_0.3.1 generics_0.0.2 xfun_0.14 R6_2.4.1
[26] doParallel_1.0.15 locfit_1.5-9.4 bitops_1.0-6 assertthat_0.2.1 promises_1.1.1
[31] scales_1.1.1 gtable_0.3.0 processx_3.4.2 rlang_0.4.6 genefilter_1.70.0
[36] scatterplot3d_0.3-41 splines_4.0.1 lazyeval_0.2.2 rstatix_0.5.0 dichromat_2.0-0
[41] broom_0.5.6 BiocManager_1.30.10 reshape2_1.4.4 abind_1.4-5 modelr_0.1.8
[46] backports_1.1.7 httpuv_1.5.4 tools_4.0.1 usethis_1.6.1 ellipsis_0.3.1
[51] gplots_3.0.3 sessioninfo_1.1.1 ggnet_0.1.0 Rcpp_1.0.4.6 zlibbioc_1.34.0
[56] RCurl_1.98-1.2 ps_1.3.3 prettyunits_1.1.1 Wrench_1.6.0 cowplot_1.0.0
[61] ggrepel_0.8.2 haven_2.3.1 cluster_2.1.0 fs_1.4.1 tinytex_0.23
[66] magrittr_1.5 data.table_1.12.8 openxlsx_4.1.5 reprex_0.3.0 mvnfast_0.2.5
[71] pkgload_1.1.0 hms_0.5.3 mime_0.9 xtable_1.8-4 klaR_0.6-15
[76] XML_3.99-0.3 rio_0.5.16 shape_1.4.4 testthat_2.3.2 compiler_4.0.1
[81] maps_3.3.0 KernSmooth_2.23-17 crayon_1.3.4 htmltools_0.4.0 mgcv_1.8-31
[86] later_1.1.0.1 geneplotter_1.66.0 lubridate_1.7.9 DBI_1.1.0 dbplyr_1.4.4
[91] MASS_7.3-51.6 ade4_1.7-15 car_3.0-8 cli_2.0.2 gdata_2.18.0
[96] igraph_1.2.5 pkgconfig_2.0.3 foreign_0.8-80 plotly_4.9.2.1 xml2_1.3.2
[101] foreach_1.5.0 annotate_1.66.0 multtest_2.44.0 XVector_0.28.0 rvest_0.3.5
[106] callr_3.4.3 digest_0.6.25 Biostrings_2.56.0 cellranger_1.1.0 curl_4.3
[111] shiny_1.4.0.2 gtools_3.8.2 lifecycle_0.2.0 nlme_3.1-148 jsonlite_1.6.1
[116] Rhdf5lib_1.10.0 network_1.16.0 carData_3.0-4 mapproj_1.2.7 viridisLite_0.3.0
[121] desc_1.2.0 fansi_0.4.1 labelled_2.4.0 pillar_1.4.4 fastmap_1.0.1
[126] httr_1.4.1 pkgbuild_1.0.8 survival_3.1-12 glue_1.4.1 remotes_2.1.1
[131] zip_2.0.4 iterators_1.0.12 bit_1.1-15.2 stringi_1.4.6 blob_1.2.1
[136] caTools_1.18.0 memoise_1.1.0
Problem:
I am performing analyses on metagenomic data in R, requiring a large number of packages to be installed and I updated to the latest versions of R and RStudio available in late May 2020. Having encountered some problems with Rtools40 not being visible, I reinstalled R today and added entries to the PATH file (as described in Rtools not found after R 4.0.0 installation). This appeared to remedy the issues I had with "the Rtools needs to be installed error". However, one of the packages I need install is the BiodiversityR package. The installation appears to proceed correctly, however when I load this package into the system library RStudio encounters and fatal error and has to restart. This is the code I have run by this stage:
#> assign working directory
setwd("F:/Work/Sequencing/R/MS2/Rdata")
#> following a new install, verify Rtools40 bin directory is included in "PATH" # NB this previously
posed a problem for any packages that needed to be compiled in R 4.0+
Sys.getenv("PATH")
#> If not, then:
Sys.setenv(PATH = paste(Sys.getenv("PATH"), "C:\\RTools40", "C:\\RTools40\\mingw64\\bin",
"C:\\RTools40\\usr\\bin",sep = ";"))
Sys.setenv(BINPREF = "C:/rtools40/mingw64/bin/")
#>install necessary packages for project
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install(c("phyloseq", "metagenomeSeq", "S4Vectors", "DESeq2"))
install.packages('ggplot2')
install.packages("colorspace")
install.packages("tidyverse")
install.packages("devtools")
devtools::install_github("grunwaldlab/metacoder")
devtools::install_github("adw96/breakaway")
devtools::install_github("adw96/DivNet")
#>Install the rest of the packages from CRAN
install.packages("agricolae")
install.packages("pals")
install.packages("Polychrome")
install.packages("forcats")
install.packages("ggpubr")
install.packages("BiodiversityR")
install.packages("glmnet")
install.packages("remotes")
remotes::install_github("MadsAlbertsen/ampvis2")
##>load libraries
library(breakaway)
library(DivNet)
library(ggpubr)
library(phyloseq)
library(metagenomeSeq)
library(S4Vectors)
library(ape)
library(ggplot2)
library(plyr)
library(dplyr)
library(biomformat)
library(DESeq2)
library(vegan)
library(metacoder)
library(taxa)
library(ggplot2)
library(dplyr)
library(readr)
library(stringr)
library(agricolae)
library(pals)
library(Polychrome)
library(forcats)
library(colorspace)
library(tidyverse)
library(readxl)
library(ampvis2)
library(BiodiversityR) #####This is where RStudio crashes
```r
When I run this last line of code the following output appears in the console before the crash
> library(BiodiversityR)
Loading required package: tcltk
Loading required package: vegan
Loading required package: permute
Loading required package: lattice
This is vegan 2.5-6
Loading required package: vegan3d
Loading required package: rgl
Registered S3 methods overwritten by 'lme4':
method from
cooks.distance.influence.merMod car
influence.merMod car
dfbeta.influence.merMod car
dfbetas.influence.merMod car
As follows
No DUMPFILE exists in the registry
Any help would be appreciated. Also note that this is the first time posting to the forum, so apologies for any inadvertent mistakes with regard to posting conventions / rules etc
Thanks in advance!